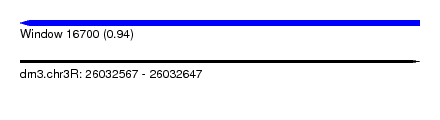
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 26,032,567 – 26,032,647 |
| Length | 80 |
| Max. P | 0.944636 |

| Location | 26,032,567 – 26,032,647 |
|---|---|
| Length | 80 |
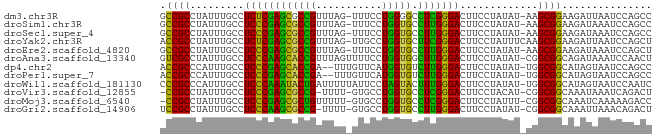
| Sequences | 12 |
| Columns | 82 |
| Reading direction | reverse |
| Mean pairwise identity | 80.45 |
| Shannon entropy | 0.40454 |
| G+C content | 0.52797 |
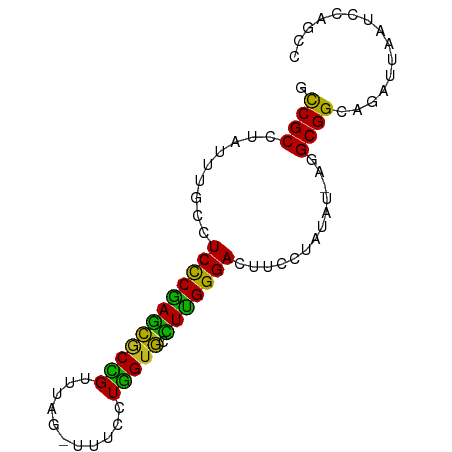
| Mean single sequence MFE | -23.52 |
| Consensus MFE | -19.85 |
| Energy contribution | -18.53 |
| Covariance contribution | -1.32 |
| Combinations/Pair | 1.62 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.944636 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
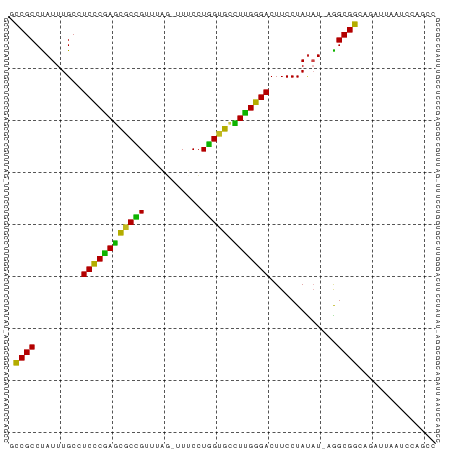
>dm3.chr3R 26032567 80 - 27905053 GCCGCCUAUUUGCCUCUCGAGCGCCGUUUAG-UUUCCUGGGGCCUCGGGACUUCCUAUAU-AAGCGGAAGAUUAAUCCAGCC .((((.(((.....(((((((.(((..((((-....))))))))))))))........))-).))))............... ( -21.52, z-score = 0.45, R) >droSim1.chr3R 25700774 80 - 27517382 GCCGCCUAUUUGCCUCCCGAGCGCCGUUUAG-UUUCCUGGUGCCUUGGGACUUCCUAUAU-AAGCGGAAGAUAAAUCCAGCC .((((.(((.....((((((((((((.....-.....))))).)))))))........))-).))))............... ( -22.52, z-score = -1.11, R) >droSec1.super_4 4884544 80 - 6179234 GCCGCCUAUUUGCCUCCCGAGCGCCGUUUAG-UUUCCUGGUGCCUUGGGACUUCCUAUAU-AAGCGGAAGAUAAAUCCAGCC .((((.(((.....((((((((((((.....-.....))))).)))))))........))-).))))............... ( -22.52, z-score = -1.11, R) >droYak2.chr3R 26965847 81 - 28832112 ACCGCCUAUUUGCCUCUCGAGCGCCGUUUAG-UUGCCUGGUGCUUCGGGACUUCCUAUUUCAAGCGGAAGAUUAAUCCAGCU ...((.........((((((((((((.....-.....)))))).))))))(((((..........))))).........)). ( -21.50, z-score = -0.31, R) >droEre2.scaffold_4820 8504746 80 + 10470090 GCCGCCUAUUUGCCUCCCGAGCGCCGUUUAG-UUUCCUGGUGCCUUGGGACUUCCUAUAU-AAGCGGAAGAUAAAUCCAGCU ...((..(((((..((((((((((((.....-.....))))).)))))))(((((.....-....))))).)))))...)). ( -22.70, z-score = -1.03, R) >droAna3.scaffold_13340 21067882 81 - 23697760 GUCGCCUAUUUGCCUCCCAAGCACCGUUUAGUUUUCCUGGUGGCUUGGGACUUCCUAUAU-CGGCGGCAGAUAAAUCCAACU ......((((((((((((((((((((...........)))).))))))))...((.....-.)).))))))))......... ( -26.70, z-score = -2.64, R) >dp4.chr2 29319311 79 + 30794189 ACCGCCCAUUUGCCUCCCGAGCACCGA--UUUGUUCAUGGUGUCUUGGGACUUCCUAUAU-UGGCGGCAUAGUAAUCCAGCC .(((((.((.....(((((((((((((--.....))..)))).)))))))........))-.)))))............... ( -24.42, z-score = -1.83, R) >droPer1.super_7 168112 79 + 4445127 ACCGCCCAUUUGCCUCCCGAGCACCGA--UUUGUUCAUGGUGUCUUGGGACUUCCUAUAU-UGGCGGCAUAGUAAUCCAGCC .(((((.((.....(((((((((((((--.....))..)))).)))))))........))-.)))))............... ( -24.42, z-score = -1.83, R) >droWil1.scaffold_181130 9497887 81 - 16660200 CCCGCCCAUUUGCCUCCCAAAUACUGAUUUUUAUUCCUAGUACUUUGGGACUUCCUAUAU-UGGCGGCAUAGUAAUCCAAUC .(((((.((.....((((((((((((...........))))).)))))))........))-.)))))............... ( -18.82, z-score = -2.35, R) >droVir3.scaffold_12855 3115926 78 - 10161210 -CCGCCUAUUUGCCUCCCGAGCGCCG-UUUU-GUGCCUGGUGCCUCGGGACUUCCUACAU-CGGCGGCAAAUAAAUCAGACU -.....((((((((((((((((((((-....-.....))))).)))))))...((.....-.)).))))))))......... ( -28.20, z-score = -2.49, R) >droMoj3.scaffold_6540 24062364 79 + 34148556 -CCGCCUAUUUGCCUCCCGAGCGCUGUUUUU-GUGCCUGGUGCCUCGGGACUUCCUAUUU-CGGCGGCAAAUCAAAAAGACC -......((((((((((((((((((((....-..))..)))).)))))))...((.....-.)).))))))).......... ( -24.40, z-score = -1.37, R) >droGri2.scaffold_14906 5811617 79 - 14172833 UCCGCCUAUUUGCCUCCCAAGCGCCG-UUUU-GUGCCUGGUGCCUUGGGACUUCCUAUAU-CGGCGGCAAAUUAAACAGACU .......(((((((((((((((((((-....-.....))))).)))))))...((.....-.)).))))))).......... ( -24.50, z-score = -1.63, R) >consensus GCCGCCUAUUUGCCUCCCGAGCGCCGUUUAG_UUUCCUGGUGCCUUGGGACUUCCUAUAU_AGGCGGCAGAUUAAUCCAGCC .((((.........((((((((((((...........))))).))))))).............))))............... (-19.85 = -18.53 + -1.32)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:56:37 2011