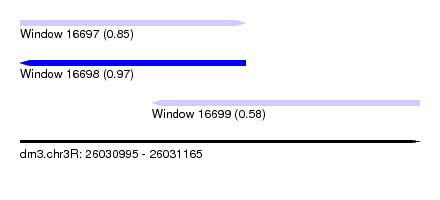
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 26,030,995 – 26,031,165 |
| Length | 170 |
| Max. P | 0.968641 |

| Location | 26,030,995 – 26,031,091 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 79.00 |
| Shannon entropy | 0.37712 |
| G+C content | 0.52585 |
| Mean single sequence MFE | -22.76 |
| Consensus MFE | -12.27 |
| Energy contribution | -12.64 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.845093 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
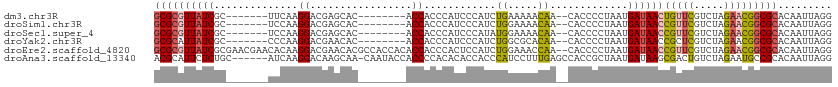
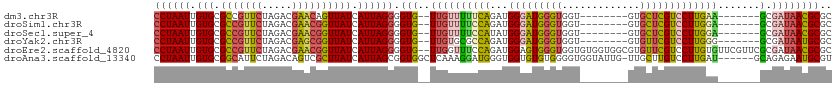
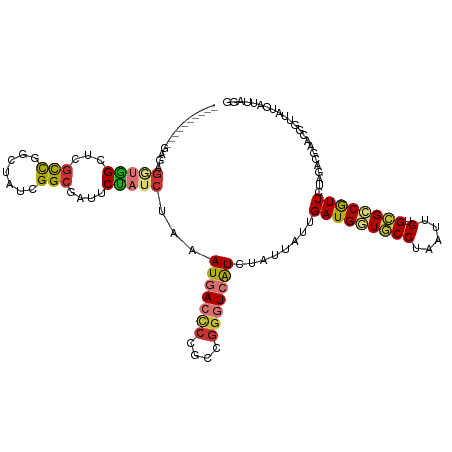
>dm3.chr3R 26030995 96 + 27905053 GCGCGUUAUCGC-------UUCAAGGACGAGCAC--------ACCACCCAUCCCAUCUGAAAAACAA--CACCCCUAAUGAUAACUGUUCGUCUAGAACGGCGCACAAUUAGG ((((((((((((-------((.......))))..--------........((......)).......--..........))))))(((((.....)))))))))......... ( -19.80, z-score = -1.41, R) >droSim1.chr3R 25699203 96 + 27517382 GCGCGUUAUCGC-------UCCAAGGACGAGCAC--------ACCACCCAUCCCAUCUGGAAAACAA--CACCCCUAAUGAUAACCGUUCGUCUAGAACGGCGCACAAUUAGG ((((((((((((-------((.......))))..--------........(((.....)))......--..........))))))(((((.....)))))))))......... ( -26.60, z-score = -3.35, R) >droSec1.super_4 4883009 96 + 6179234 GCGCGUUAUCGC-------UCCAAGGACGAGCAC--------ACCACCCAUCCCAUAUGGAAAACAA--CACCCCUAAUGAUAACCGUUCGUCUAGAACGGCGCACAAUUAGG ((((((((((((-------((.......))))..--------.....((((.....)))).......--..........))))))(((((.....)))))))))......... ( -27.40, z-score = -3.75, R) >droYak2.chr3R 26964252 96 + 28832112 GCGCAUUAUCGC-------CCCAAGGACGAACAC--------ACCACCCAUCCCAUCUGGCGCACAA--CACCCCUAAUGAUAACCGCUCGUCUAGAACGGCGCACAAUUAGG ((((....((((-------(....)).)))....--------......((.......))))))....--....((((((......(((.(((.....))))))....)))))) ( -20.20, z-score = -1.39, R) >droEre2.scaffold_4820 8503168 111 - 10470090 GCGCGUUAUCGCGAACGAACACAAGGACGAACACGCCACCACACCACCCACUCCAUCUGGAAACCAA--CACCCCUAAUGAUAACCGUUCGUCUAGAACGGCGCACAAUUAGG (((((((((((.............((.((....))))..............(((....)))......--.........)))))))(((((.....)))))))))......... ( -25.20, z-score = -2.24, R) >droAna3.scaffold_13340 21066021 106 + 23697760 ACGCAUUCUCUGC------AUCAAGGACAAGCAA-CAAUACCACCCCACACACCACCCAUCCUUUGAGCCACCGCUAAUGAUAAGCGACUGUCUAGAAUGCCGCACAAUUAGG ..(((((((..((------(((((((........-..........................)))))).....((((.......))))..)))..)))))))............ ( -17.38, z-score = -0.85, R) >consensus GCGCGUUAUCGC_______UCCAAGGACGAGCAC________ACCACCCAUCCCAUCUGGAAAACAA__CACCCCUAAUGAUAACCGUUCGUCUAGAACGGCGCACAAUUAGG ((((((((((..............((..........................))...((.....)).............))))))(((((.....)))))))))......... (-12.27 = -12.64 + 0.37)
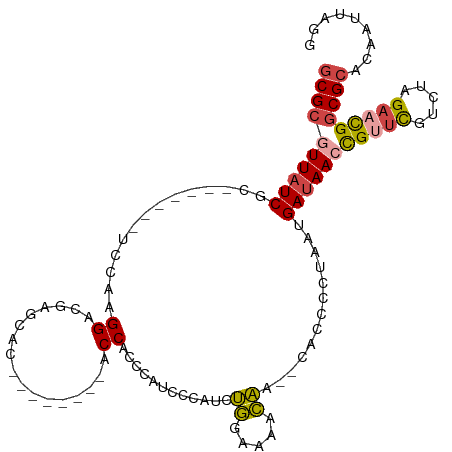
| Location | 26,030,995 – 26,031,091 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 79.00 |
| Shannon entropy | 0.37712 |
| G+C content | 0.52585 |
| Mean single sequence MFE | -37.75 |
| Consensus MFE | -20.09 |
| Energy contribution | -20.46 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.968641 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
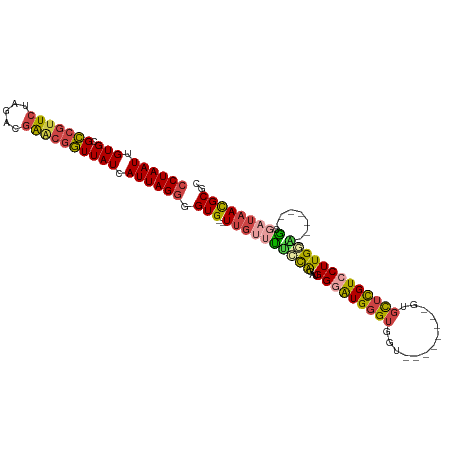
>dm3.chr3R 26030995 96 - 27905053 CCUAAUUGUGCGCCGUUCUAGACGAACAGUUAUCAUUAGGGGUG--UUGUUUUUCAGAUGGGAUGGGUGGU--------GUGCUCGUCCUUGAA-------GCGAUAACGCGC ((((((.(((.((.((((.....)))).))))).)))))).(((--((((((((((...(((((((((...--------..)))))))))))))-------).)))))))).. ( -33.50, z-score = -2.41, R) >droSim1.chr3R 25699203 96 - 27517382 CCUAAUUGUGCGCCGUUCUAGACGAACGGUUAUCAUUAGGGGUG--UUGUUUUCCAGAUGGGAUGGGUGGU--------GUGCUCGUCCUUGGA-------GCGAUAACGCGC ((((((.(((.(((((((.....)))))))))).)))))).(((--((((((((((...(((((((((...--------..)))))))))))))-------).)))))))).. ( -42.00, z-score = -4.38, R) >droSec1.super_4 4883009 96 - 6179234 CCUAAUUGUGCGCCGUUCUAGACGAACGGUUAUCAUUAGGGGUG--UUGUUUUCCAUAUGGGAUGGGUGGU--------GUGCUCGUCCUUGGA-------GCGAUAACGCGC ((((((.(((.(((((((.....)))))))))).)))))).(((--((((((((((...(((((((((...--------..)))))))))))))-------).)))))))).. ( -42.00, z-score = -4.53, R) >droYak2.chr3R 26964252 96 - 28832112 CCUAAUUGUGCGCCGUUCUAGACGAGCGGUUAUCAUUAGGGGUG--UUGUGCGCCAGAUGGGAUGGGUGGU--------GUGUUCGUCCUUGGG-------GCGAUAAUGCGC ((((((.(((.(((((((.....)))))))))).)))))).(((--((((.((((....((((((..(...--------..)..))))))...)-------)))))))))).. ( -40.40, z-score = -3.34, R) >droEre2.scaffold_4820 8503168 111 + 10470090 CCUAAUUGUGCGCCGUUCUAGACGAACGGUUAUCAUUAGGGGUG--UUGGUUUCCAGAUGGAGUGGGUGGUGUGGUGGCGUGUUCGUCCUUGUGUUCGUUCGCGAUAACGCGC ((((((.(((.(((((((.....)))))))))).)))))).(((--(((...(((....)))(((..(((...((.((((....)))))).....)))..)))..)))))).. ( -37.40, z-score = -1.25, R) >droAna3.scaffold_13340 21066021 106 - 23697760 CCUAAUUGUGCGGCAUUCUAGACAGUCGCUUAUCAUUAGCGGUGGCUCAAAGGAUGGGUGGUGUGUGGGGUGGUAUUG-UUGCUUGUCCUUGAU------GCAGAGAAUGCGU ............(((((((...((.(((((.......)))))))((...(((((..((..(...(((......)))..-)..))..)))))...------))..))))))).. ( -31.20, z-score = -0.23, R) >consensus CCUAAUUGUGCGCCGUUCUAGACGAACGGUUAUCAUUAGGGGUG__UUGUUUUCCAGAUGGGAUGGGUGGU________GUGCUCGUCCUUGGA_______GCGAUAACGCGC ((((((.(((.(((((((.....)))))))))).)))))).(((..((((.........((((((((...............))))))))...........))))...))).. (-20.09 = -20.46 + 0.37)
| Location | 26,031,051 – 26,031,165 |
|---|---|
| Length | 114 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.94 |
| Shannon entropy | 0.51123 |
| G+C content | 0.49098 |
| Mean single sequence MFE | -34.80 |
| Consensus MFE | -17.76 |
| Energy contribution | -17.90 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.584008 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
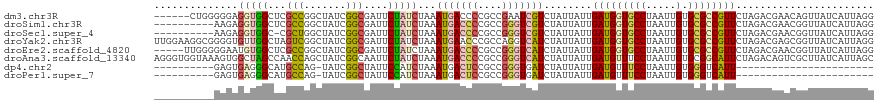
>dm3.chr3R 26031051 114 - 27905053 ------CUGGGGGAGGUGGCUCGCCGGCUAUCGGCGAUUCUAUCUAAAUGACCCCGCCGAAUCGUCUAUUAUUGAUGGUGCCUAAUUGUGCGCCGUUCUAGACGAACAGUUAUCAUUAGG ------(((((((((((((.((((((.....))))))..))))))......))))......(((((((.....(((((((((.....).)))))))).))))))).)))........... ( -41.90, z-score = -2.34, R) >droSim1.chr3R 25699259 110 - 27517382 ----------AAGAGGUGGCUCGCCGGCUAUCGGCGAUUCUAUCUAAAUGACCCCGCCGGGUCGUCUAUUAUUGAUGGUGCCUAAUUGUGCGCCGUUCUAGACGAACGGUUAUCAUUAGG ----------...((((((.((((((.....))))))..)))))).(((((....((((..(((((((.....(((((((((.....).)))))))).))))))).))))..)))))... ( -42.80, z-score = -2.90, R) >droSec1.super_4 4883065 109 - 6179234 ----------AAGAGGUGGC-CGCUGGCUAUCGGCGAUUCUAUCUAAAUGACCCCGCCGGGUCGUCUAUUAUUGAUGGUGCCUAAUUGUGCGCCGUUCUAGACGAACGGUUAUCAUUAGG ----------...((((((.-(((((.....)))))...)))))).(((((....((((..(((((((.....(((((((((.....).)))))))).))))))).))))..)))))... ( -38.50, z-score = -1.82, R) >droYak2.chr3R 26964308 120 - 28832112 UUGGAAGGCGGGGUGUUGGCUAGUCGGCUAUCGGCGAUUCUAUCUAAAUGAACCCGCCAGGUCAUCUAUUAUUGAUGGUGCCUAAUUGUGCGCCGUUCUAGACGAGCGGUUAUCAUUAGG ......((((((...((((.((((((.(....).)))..))).)))).....))))))...(((((.......)))))..((((((.(((.(((((((.....)))))))))).)))))) ( -40.80, z-score = -1.39, R) >droEre2.scaffold_4820 8503239 115 + 10470090 -----UUGGGGGAAUGUGGCUCGCCGGCUAUCGGCGAUUCUAUCUAAAUGACCCCGCCGGGUCAUCUAUUAUUGAUGGUGCCUAAUUGUGCGCCGUUCUAGACGAACGGUUAUCAUUAGG -----..((((..((((((.((((((.....))))))..))))....))..))))(((..((((........))))))).((((((.(((.(((((((.....)))))))))).)))))) ( -40.20, z-score = -1.50, R) >droAna3.scaffold_13340 21066087 120 - 23697760 AGGGUGGUAAAGUGGCUAGCCAACCAGCUAUCGGCAAUUCUAUCUAAAUGACCCCGCCGGGUCAUCUAUUAUUGAUGUUUCCUAAUUGUGCGGCAUUCUAGACAGUCGCUUAUCAUUAGC ..((((((((.((((((.(((...........))).......((((((((.((.(((.(((.((((.......))))...)))....))).)))))).)))).))))))))))))))... ( -32.80, z-score = -0.78, R) >dp4.chr2 29317689 86 + 30794189 ----------GAGUGAGGGCAUGCCAG-UAUCGGCUAUUCCAUCUAAAUGACUCCGCCGGGUGAUCUAUUAUUGAUGUUUCCUAAUUGUGGGUCAUU----------------------- ----------......(((...(((..-....)))...))).....((((((.((((.(((..(((.......)))....)))....))))))))))----------------------- ( -20.70, z-score = 0.37, R) >droPer1.super_7 166484 86 + 4445127 ----------GAGUGAGGGCAUGCCAG-UAUCGGCUAUUCCAUCUAAAUGACUCCGCCGGGUGAUCUAUUAUUGAUGUUUCCUAAUUGUGGGUCAUU----------------------- ----------......(((...(((..-....)))...))).....((((((.((((.(((..(((.......)))....)))....))))))))))----------------------- ( -20.70, z-score = 0.37, R) >consensus __________GAGAGGUGGCUCGCCGGCUAUCGGCGAUUCUAUCUAAAUGACCCCGCCGGGUCAUCUAUUAUUGAUGGUGCCUAAUUGUGCGCCGUUCUAGACGAACGGUUAUCAUUAGG ..............(((((...(((.......)))....)))))...(((((((....)))))))........(((((((((.....).))))))))....................... (-17.76 = -17.90 + 0.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:56:36 2011