
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 26,022,190 – 26,022,309 |
| Length | 119 |
| Max. P | 0.970404 |

| Location | 26,022,190 – 26,022,294 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 91.63 |
| Shannon entropy | 0.13533 |
| G+C content | 0.46538 |
| Mean single sequence MFE | -20.55 |
| Consensus MFE | -18.95 |
| Energy contribution | -19.08 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.738721 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
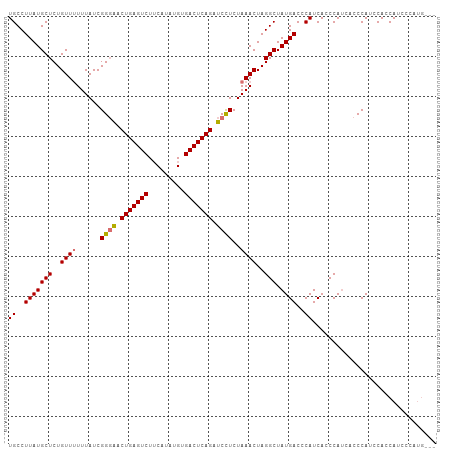
>dm3.chr3R 26022190 104 - 27905053 UGCCUUAUGCUCUGUUUUUUAUCGGGACCUGAGUCUUCAUAUGUGACUCAGAUCCUCUAAACUAGGCUAUGACCCAUCACCCAACGCCCAUCCACCAUCCCAUG--- ((..(((((((..((((......((((.(((((((.........))))))).))))..))))..))).))))..))............................--- ( -21.70, z-score = -1.95, R) >droEre2.scaffold_4820 8494216 107 + 10470090 UGCCUUAUGCUCUGUUGUUUAUCGGGGACUGAGUCUGCAUAUGUGACUCAGAUCUUCUAAACUAGGCUAUGACCCAUCACCCAUCACCCAUCCAACAUCCAACAUCC .((((...........(((((..((((((((((((.((....))))))))).)))))))))).))))........................................ ( -21.50, z-score = -0.28, R) >droSec1.super_4 4874680 104 - 6179234 UGCCUUAUGCUCUGUUUUUUAUCGGGAACUGAGUCUUCAUAUGUGACUCAGAUCCUCUAAACUAGGCUAUGACACAUCACCCAUCACCCAUCCACCAUCCCAUG--- ((..(((((((..((((......((((.(((((((.........))))))).))))..))))..))).))))..))............................--- ( -21.70, z-score = -1.89, R) >droSim1.chr3R 25689983 104 - 27517382 UGCCUUAUGCUCUGUUUUUUAUCGGCAACUGAGUCUUCAUAUGUGACUCAGAUCCUCUAAACUAGGCUAUGACCCAUCACCCAUACCCCAUCCACCAUCCCAUG--- .((((..((((............)))).(((((((.........)))))))............)))).....................................--- ( -17.30, z-score = -1.39, R) >consensus UGCCUUAUGCUCUGUUUUUUAUCGGGAACUGAGUCUUCAUAUGUGACUCAGAUCCUCUAAACUAGGCUAUGACCCAUCACCCAUCACCCAUCCACCAUCCCAUG___ ((..(((((((..((((......((((.(((((((.........))))))).))))..))))..))).))))..))............................... (-18.95 = -19.08 + 0.12)


| Location | 26,022,219 – 26,022,309 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 74.47 |
| Shannon entropy | 0.49281 |
| G+C content | 0.44934 |
| Mean single sequence MFE | -26.12 |
| Consensus MFE | -14.95 |
| Energy contribution | -14.82 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.970404 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
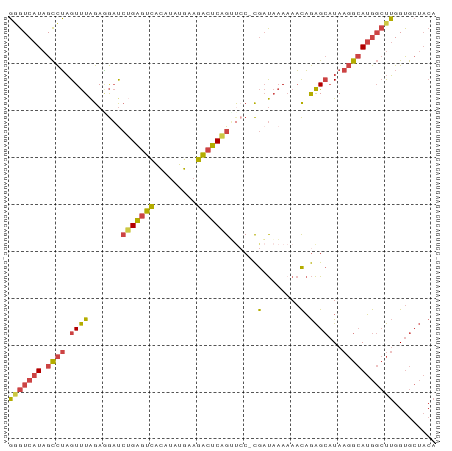
>dm3.chr3R 26022219 90 + 27905053 GGGUCAUAGCCUAGUUUAGAGGAUCUGAGUCACAUAUGAAGACUCAGGUCC-CGAUAAAAAACAGAGCAUAAGGCAUGGCUUGGUGCUACA (((((((.((((.((((.(.(((((((((((.........)))))))))))-)(........).))))...)))))))))))......... ( -29.90, z-score = -2.86, R) >droEre2.scaffold_4820 8494248 90 - 10470090 GGGUCAUAGCCUAGUUUAGAAGAUCUGAGUCACAUAUGCAGACUCAGUCCC-CGAUAAACAACAGAGCAUAAGGCAUGGCUUGGUGCUACA ((((....)))).(((((...((.(((((((.........)))))))))..-...))))).....(((((.((((...)))).)))))... ( -24.50, z-score = -0.98, R) >droYak2.chr3R 26955982 91 + 28832112 GGGUCAUAGCCUAGUUUAGAAGACCGGAGUCACAUAUGAAGACUCCGGUCCUCGAUAAAAAACAGAGCAUAAGGCAUGGCUUGGUGCUACA (((((((.((((.........((((((((((.........))))))))))(((...........)))....)))))))))))......... ( -32.60, z-score = -3.64, R) >droSec1.super_4 4874709 90 + 6179234 GUGUCAUAGCCUAGUUUAGAGGAUCUGAGUCACAUAUGAAGACUCAGUUCC-CGAUAAAAAACAGAGCAUAAGGCAUGGCUUGGUGCUACA (..(((.((((...((((..(((.(((((((.........))))))).)))-...)))).......((.....))..)))))))..).... ( -26.70, z-score = -2.18, R) >droSim1.chr3R 25690012 90 + 27517382 GGGUCAUAGCCUAGUUUAGAGGAUCUGAGUCACAUAUGAAGACUCAGUUGC-CGAUAAAAAACAGAGCAUAAGGCAUGGCUUGGUGCUACA ((((....))))..((((..((..(((((((.........)))))))...)-)..))))......(((((.((((...)))).)))))... ( -26.00, z-score = -1.73, R) >droVir3.scaffold_12855 3095439 85 + 10161210 UGGUCAU-GUUGUGUCUGGAUAUUAGGUAUUUUGGGAGUGGGAGCGGACGCUUAUCUUAAACUUAGGAUUAACAAAAACACAGGCG----- ..(((.(-(((.(((...(((.((((((...((((((....((((....)))).)))))))))))).))).)))..))))..))).----- ( -17.00, z-score = -0.30, R) >consensus GGGUCAUAGCCUAGUUUAGAGGAUCUGAGUCACAUAUGAAGACUCAGUUCC_CGAUAAAAAACAGAGCAUAAGGCAUGGCUUGGUGCUACA (((((((.((((.((((.......(((((((.........)))))))......(........).))))...)))))))))))......... (-14.95 = -14.82 + -0.13)


| Location | 26,022,219 – 26,022,309 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 74.47 |
| Shannon entropy | 0.49281 |
| G+C content | 0.44934 |
| Mean single sequence MFE | -21.72 |
| Consensus MFE | -10.81 |
| Energy contribution | -12.53 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.772652 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
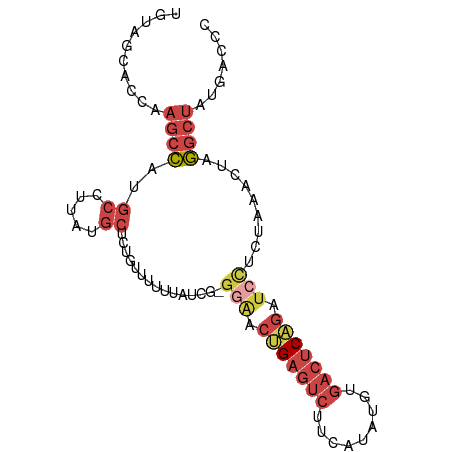
>dm3.chr3R 26022219 90 - 27905053 UGUAGCACCAAGCCAUGCCUUAUGCUCUGUUUUUUAUCG-GGACCUGAGUCUUCAUAUGUGACUCAGAUCCUCUAAACUAGGCUAUGACCC .(((((...((((...((.....))...))))......(-(((.(((((((.........))))))).)))).........)))))..... ( -22.40, z-score = -1.47, R) >droEre2.scaffold_4820 8494248 90 + 10470090 UGUAGCACCAAGCCAUGCCUUAUGCUCUGUUGUUUAUCG-GGGACUGAGUCUGCAUAUGUGACUCAGAUCUUCUAAACUAGGCUAUGACCC .(((((....(((...((.....))...)))(((((..(-(((((((((((.((....))))))))).))))))))))...)))))..... ( -24.30, z-score = -0.93, R) >droYak2.chr3R 26955982 91 - 28832112 UGUAGCACCAAGCCAUGCCUUAUGCUCUGUUUUUUAUCGAGGACCGGAGUCUUCAUAUGUGACUCCGGUCUUCUAAACUAGGCUAUGACCC .(((((...((((...((.....))...))))......(((((((((((((.........)))))))))))))........)))))..... ( -31.80, z-score = -4.06, R) >droSec1.super_4 4874709 90 - 6179234 UGUAGCACCAAGCCAUGCCUUAUGCUCUGUUUUUUAUCG-GGAACUGAGUCUUCAUAUGUGACUCAGAUCCUCUAAACUAGGCUAUGACAC .(((((...((((...((.....))...))))......(-(((.(((((((.........))))))).)))).........)))))..... ( -22.40, z-score = -1.42, R) >droSim1.chr3R 25690012 90 - 27517382 UGUAGCACCAAGCCAUGCCUUAUGCUCUGUUUUUUAUCG-GCAACUGAGUCUUCAUAUGUGACUCAGAUCCUCUAAACUAGGCUAUGACCC ........((((((.((((...................)-))).(((((((.........))))))).............)))).)).... ( -20.81, z-score = -1.39, R) >droVir3.scaffold_12855 3095439 85 - 10161210 -----CGCCUGUGUUUUUGUUAAUCCUAAGUUUAAGAUAAGCGUCCGCUCCCACUCCCAAAAUACCUAAUAUCCAGACACAAC-AUGACCA -----....(((((((.((((((((.((....)).))).(((....))).................)))))...)))))))..-....... ( -8.60, z-score = -0.41, R) >consensus UGUAGCACCAAGCCAUGCCUUAUGCUCUGUUUUUUAUCG_GGAACUGAGUCUUCAUAUGUGACUCAGAUCCUCUAAACUAGGCUAUGACCC ..........((((..((.....))...............(((.(((((((.........))))))).))).........))))....... (-10.81 = -12.53 + 1.72)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:56:31 2011