
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 26,019,862 – 26,019,955 |
| Length | 93 |
| Max. P | 0.514592 |

| Location | 26,019,862 – 26,019,955 |
|---|---|
| Length | 93 |
| Sequences | 7 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 64.45 |
| Shannon entropy | 0.60803 |
| G+C content | 0.43032 |
| Mean single sequence MFE | -21.27 |
| Consensus MFE | -8.94 |
| Energy contribution | -9.67 |
| Covariance contribution | 0.74 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.514592 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 26019862 93 + 27905053 ------------------AACAUA--CGCAUAUGUAAAUUUAUCCCGAUAUCCAACCGAUUCGAGCAUGGGAAAGCAAACGAACCUAUUGCCAAAUG-------AUUUGGCAUCUGCAAA ------------------......--.(((............(((((...((..........))...)))))................((((((...-------..))))))..)))... ( -15.40, z-score = 0.13, R) >droSim1.chr3R 25687701 111 + 27517382 AACAUACGCACAUAUGUGUAUGUA--UGUAUAUGUAAAUUUAACCCGAUACCCAACCGAUUCGAUCAUGGGAAAGCAAACGAACCCAUUGCCAAAUG-------AUUCGGCAUCUGCAAA .(((((((((....))))))))).--((((.((((...............((((...(((...))).))))........((((..((((....))))-------.)))))))).)))).. ( -25.20, z-score = -1.44, R) >droSec1.super_4 4872529 110 + 6179234 ACCAUACGCACAUAUGCGUAUGUA--UUUAUAUGUAAAUUUAUCCCGAUACCCAACCGAUUCGAUCAUGGGAA-GCAAACAAACCCAUUGCCAAAUG-------AUUCGGCAUCUGCAAA ..((((((((....))))))))..--......((((.........(((............)))...(((((..-.........)))))((((.....-------....))))..)))).. ( -20.60, z-score = -0.36, R) >droEre2.scaffold_4820 8491921 113 - 10470090 AACACACGCACAUAUGUAUAUUUAAAUGUAUAUGCAGACCAAUCACGAUAGCCAGCCGAUUUGAUCAUGGGAAAGCAAACGAACCCAUUGCCAAAUG-------AUUUGGCAUCUGAAGA .......(((.((((((........)))))).))).......(((.(((.(((((...(((((...(((((...(....)...)))))...))))).-------..)))))))))))... ( -23.50, z-score = -1.02, R) >droAna3.scaffold_13340 21054429 78 + 23697760 ----------------------------------CCGAUCCGCUUCGAU-CCCGUCCGAACCGAGCAUGGGAAAGCAAACGCACCUAUUGCCAAACG-------AAUUGGCAUCUGUAAA ----------------------------------.......((((...(-(((((.(.......).))))))))))....(((.....((((((...-------..))))))..)))... ( -18.40, z-score = -0.68, R) >dp4.chr2 29307488 85 - 30794189 ----------------------------------AAGCGCUAUCCCUAU-CCUAUCCGAUACGAUCAUGGGAAAGCAAACGAGCCGAUUGCCAAAUGCCAAUUUAUUUGGCAUCCGUAAA ----------------------------------....(((.((((.((-(.(((...))).)))...)))).)))........((..(((((((((......)))))))))..)).... ( -22.90, z-score = -2.33, R) >droPer1.super_7 155581 85 - 4445127 ----------------------------------AAGCGCUAUCCCUAU-CCUAUCCGAUACGAUCAUGGGAAAGCAAACGAGCCGAUUGCCAAAUGCCAAUUUAUUUGGCAUCCGUAAA ----------------------------------....(((.((((.((-(.(((...))).)))...)))).)))........((..(((((((((......)))))))))..)).... ( -22.90, z-score = -2.33, R) >consensus ___________________A__UA_____AUAUGUAGAUCUAUCCCGAUACCCAACCGAUUCGAUCAUGGGAAAGCAAACGAACCCAUUGCCAAAUG_______AUUUGGCAUCUGCAAA ..................................................................(((((...(....)...)))))((((((((........))))))))........ ( -8.94 = -9.67 + 0.74)

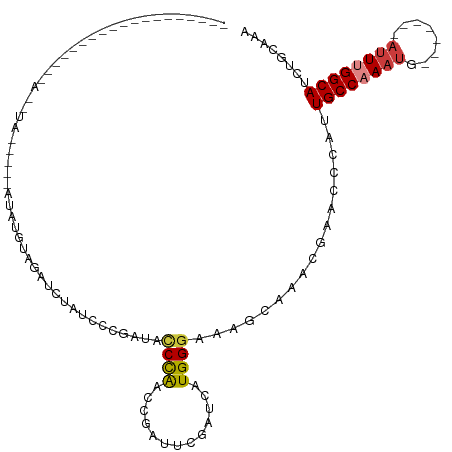
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:56:29 2011