
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 26,012,888 – 26,012,980 |
| Length | 92 |
| Max. P | 0.986877 |

| Location | 26,012,888 – 26,012,980 |
|---|---|
| Length | 92 |
| Sequences | 11 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 75.40 |
| Shannon entropy | 0.48368 |
| G+C content | 0.34935 |
| Mean single sequence MFE | -23.38 |
| Consensus MFE | -12.29 |
| Energy contribution | -12.61 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.959488 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
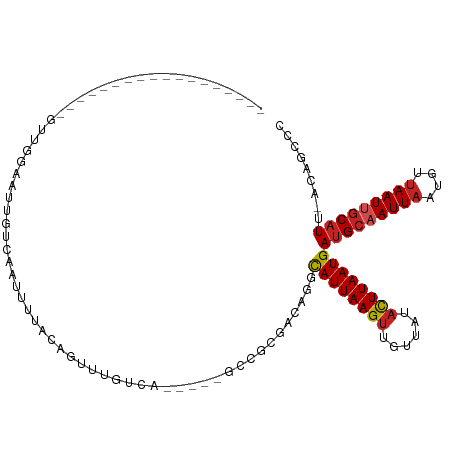
>dm3.chr3R 26012888 92 + 27905053 ------------------GUUGGAAUUGUCAAUUUUACAGUUUGUCA-----GCCGCGACAGGCAUUAAGUUGUUAUACUUAAUGAUGCAAUUAAUGUUAAUUGCAUU-AUAGCCC ------------------....(((((((.......)))))))(((.-----.....))).(((..(((((......)))))((((((((((((....))))))))))-)).))). ( -24.50, z-score = -2.64, R) >droPer1.super_7 148833 95 - 4445127 ---------------GUUUUCGGAAUUGUCAAUUUUACAGUUUGUCA-----GUCGCGAAAGGUAUUAAGUUGUUAUACUUAAUGAUGCAAUUAAUGUAAAUGGCAUU-ACAGCGG ---------------.((((((..((((.(((.........))).))-----))..)))))).((((((((......)))))))).(((...((((((.....)))))-)..))). ( -19.60, z-score = -1.17, R) >dp4.chr2 29300700 95 - 30794189 ---------------GUUUUCGGAAUUGUCAAUUUUACAGUUUGUCA-----GUCGCGAAAGGUAUUAAGUUGUUAUACUUAAUGAUGCAAUUAAUGUUAAUGGCAUU-ACAGCGG ---------------.((((((..((((.(((.........))).))-----))..)))))).((((((((......)))))))).(((...((((((.....)))))-)..))). ( -19.60, z-score = -1.18, R) >droAna3.scaffold_13340 21047542 92 + 23697760 ------------------GUUGGUAUUGUCAAUUUUACAGUUUUUCA-----GCCGCAACAGGCAUUAAGUUGUUAUACUUAAUGAUGCAAUUAAUGUUAAUUGCAUU-ACAGCCC ------------------(((((.(((((.......)))))...)))-----)).......((((((((((......)))))))((((((((((....))))))))))-...))). ( -23.90, z-score = -2.77, R) >droEre2.scaffold_4820 8484970 92 - 10470090 ------------------GUUGGAAUUGUCAAUUUUACAGUUUGUCA-----GCCGCGACAGGCAUUAAGUUGUUAUACUUAAUGAUGCAAUUAAUGUUAAUUGCAUU-ACAGCCC ------------------((((.................(((((((.-----.....)))))))(((((((......)))))))((((((((((....))))))))))-.)))).. ( -24.80, z-score = -2.61, R) >droYak2.chr3R 26946402 92 + 28832112 ------------------GUUGGAAUUGUCAAUUUUACAGUUUGUCA-----GCCGCGACAGGCAUUAAGUUGUUAUACUUAAUGAUGCAAUUAAUGUUAAUUGCAUU-ACAGCCC ------------------((((.................(((((((.-----.....)))))))(((((((......)))))))((((((((((....))))))))))-.)))).. ( -24.80, z-score = -2.61, R) >droSec1.super_4 4865644 92 + 6179234 ------------------GUUGGAAUUGUCACUUUUACAGUUUGUCA-----GCCGCGACAGGCAUUAAGUUGUUAUACUUAAUGAUGCAAUUAAUGUUAAUUGCAUU-ACAGCCC ------------------((((.................(((((((.-----.....)))))))(((((((......)))))))((((((((((....))))))))))-.)))).. ( -24.80, z-score = -2.48, R) >droSim1.chr3R 25680346 92 + 27517382 ------------------GUUGGAAUUGUCAAUUUUACAGUUUGUCA-----GCCGCGACAGGCAUUAAGUUGUUAUACUUAAUGAUGCAAUUAAUGUUAAUUGCAUU-ACAGCCC ------------------((((.................(((((((.-----.....)))))))(((((((......)))))))((((((((((....))))))))))-.)))).. ( -24.80, z-score = -2.61, R) >droWil1.scaffold_181130 9473999 78 + 16660200 ---------------------------GUUCGUAUUGUUGCUU--CA-----AC----AAAAGCAUUAAGUUGUUAUAUUUAAUGAUGCAAUUAAUGUUAAUUGCAUUUACAACCC ---------------------------(((.(((((((((...--))-----))----))...((((((((......))))))))(((((((((....))))))))).)))))).. ( -16.70, z-score = -2.20, R) >droVir3.scaffold_12855 3080861 113 + 10161210 UGUUGUUGUCGCCGCCUUUUGUCAAUUUACAAUUUCGGCACAAGUCGCGCGGUGACAAAAAGGCAUUAAGCUGUUGUACUUAAUGACGCAAUUAAUGUUAAUUACAUU-ACGAC-- .((((((((((((((...((((......))))...((((....)))).)))))))))....((((((((..(((..(.....)..)))...)))))))).........-)))))-- ( -30.10, z-score = -1.44, R) >droGri2.scaffold_14906 5784838 106 + 14172833 -------GUUGACGGCUUUUGUUCAUUUACAGUUUCGGCGAUAGCGACAAAAAAAGAAAAAGGCAUUAAGUUGUCGUACUUAAUGAUGCAAUUAAUGCUAAUUGCAUU-ACGAC-- -------(((((((((((.((((..(((....(((..(((....)).)..)))...)))..))))..))))))))).))....(((((((((((....))))))))))-)....-- ( -23.60, z-score = -0.98, R) >consensus __________________GUUGGAAUUGUCAAUUUUACAGUUUGUCA_____GCCGCGACAGGCAUUAAGUUGUUAUACUUAAUGAUGCAAUUAAUGUUAAUUGCAUU_ACAGCCC ...............................................................((((((((......))))))))(((((((((....)))))))))......... (-12.29 = -12.61 + 0.32)

| Location | 26,012,888 – 26,012,980 |
|---|---|
| Length | 92 |
| Sequences | 11 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 75.40 |
| Shannon entropy | 0.48368 |
| G+C content | 0.34935 |
| Mean single sequence MFE | -20.31 |
| Consensus MFE | -11.52 |
| Energy contribution | -11.82 |
| Covariance contribution | 0.30 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.26 |
| SVM RNA-class probability | 0.986877 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
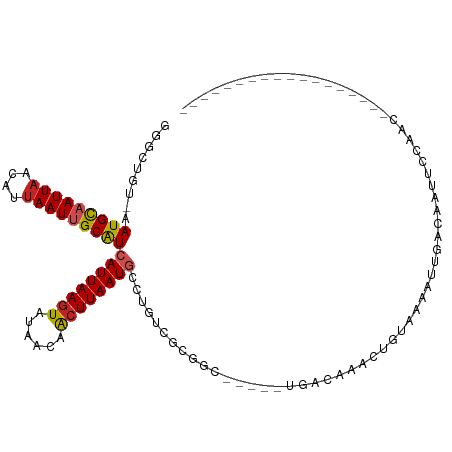
>dm3.chr3R 26012888 92 - 27905053 GGGCUAU-AAUGCAAUUAACAUUAAUUGCAUCAUUAAGUAUAACAACUUAAUGCCUGUCGCGGC-----UGACAAACUGUAAAAUUGACAAUUCCAAC------------------ ((((...-.(((((((((....))))))))).(((((((......)))))))))))(((((((.-----.......))))......))).........------------------ ( -20.10, z-score = -2.48, R) >droPer1.super_7 148833 95 + 4445127 CCGCUGU-AAUGCCAUUUACAUUAAUUGCAUCAUUAAGUAUAACAACUUAAUACCUUUCGCGAC-----UGACAAACUGUAAAAUUGACAAUUCCGAAAAC--------------- ..((..(-((((.......)))))...))...(((((((......)))))))...(((((.((.-----((.(((.........))).)).)).)))))..--------------- ( -12.60, z-score = -1.39, R) >dp4.chr2 29300700 95 + 30794189 CCGCUGU-AAUGCCAUUAACAUUAAUUGCAUCAUUAAGUAUAACAACUUAAUACCUUUCGCGAC-----UGACAAACUGUAAAAUUGACAAUUCCGAAAAC--------------- ..((..(-((((.......)))))...))...(((((((......)))))))...(((((.((.-----((.(((.........))).)).)).)))))..--------------- ( -12.60, z-score = -1.45, R) >droAna3.scaffold_13340 21047542 92 - 23697760 GGGCUGU-AAUGCAAUUAACAUUAAUUGCAUCAUUAAGUAUAACAACUUAAUGCCUGUUGCGGC-----UGAAAAACUGUAAAAUUGACAAUACCAAC------------------ (((((((-((((((((((....)))))))..((((((((......))))))))...))))))))-----........(((.......)))...))...------------------ ( -23.60, z-score = -3.30, R) >droEre2.scaffold_4820 8484970 92 + 10470090 GGGCUGU-AAUGCAAUUAACAUUAAUUGCAUCAUUAAGUAUAACAACUUAAUGCCUGUCGCGGC-----UGACAAACUGUAAAAUUGACAAUUCCAAC------------------ .((((((-.(((((((((....)))))))))((((((((......))))))))......)))))-----)............................------------------ ( -21.70, z-score = -2.57, R) >droYak2.chr3R 26946402 92 - 28832112 GGGCUGU-AAUGCAAUUAACAUUAAUUGCAUCAUUAAGUAUAACAACUUAAUGCCUGUCGCGGC-----UGACAAACUGUAAAAUUGACAAUUCCAAC------------------ .((((((-.(((((((((....)))))))))((((((((......))))))))......)))))-----)............................------------------ ( -21.70, z-score = -2.57, R) >droSec1.super_4 4865644 92 - 6179234 GGGCUGU-AAUGCAAUUAACAUUAAUUGCAUCAUUAAGUAUAACAACUUAAUGCCUGUCGCGGC-----UGACAAACUGUAAAAGUGACAAUUCCAAC------------------ ((((...-.(((((((((....))))))))).(((((((......)))))))))))(((((...-----..((.....))....))))).........------------------ ( -24.40, z-score = -3.15, R) >droSim1.chr3R 25680346 92 - 27517382 GGGCUGU-AAUGCAAUUAACAUUAAUUGCAUCAUUAAGUAUAACAACUUAAUGCCUGUCGCGGC-----UGACAAACUGUAAAAUUGACAAUUCCAAC------------------ .((((((-.(((((((((....)))))))))((((((((......))))))))......)))))-----)............................------------------ ( -21.70, z-score = -2.57, R) >droWil1.scaffold_181130 9473999 78 - 16660200 GGGUUGUAAAUGCAAUUAACAUUAAUUGCAUCAUUAAAUAUAACAACUUAAUGCUUUU----GU-----UG--AAGCAACAAUACGAAC--------------------------- (..((((..(((((((((....)))))))))....................((((((.----..-----.)--)))))))))..)....--------------------------- ( -16.40, z-score = -2.48, R) >droVir3.scaffold_12855 3080861 113 - 10161210 --GUCGU-AAUGUAAUUAACAUUAAUUGCGUCAUUAAGUACAACAGCUUAAUGCCUUUUUGUCACCGCGCGACUUGUGCCGAAAUUGUAAAUUGACAAAAGGCGGCGACAACAACA --(((((-.(((((((((....))))))))).(((((((......)))))))(((((((.(((((.((((.....)))).)...........))))))))))).)))))....... ( -31.40, z-score = -2.28, R) >droGri2.scaffold_14906 5784838 106 - 14172833 --GUCGU-AAUGCAAUUAGCAUUAAUUGCAUCAUUAAGUACGACAACUUAAUGCCUUUUUCUUUUUUUGUCGCUAUCGCCGAAACUGUAAAUGAACAAAAGCCGUCAAC------- --(((((-((((((((((....))))))))).......))))))......(((.(((((((.(((....((((....).)))......))).))..))))).)))....------- ( -17.21, z-score = -0.36, R) >consensus GGGCUGU_AAUGCAAUUAACAUUAAUUGCAUCAUUAAGUAUAACAACUUAAUGCCUGUCGCGGC_____UGACAAACUGUAAAAUUGACAAUUCCAAC__________________ .........(((((((((....)))))))))((((((((......))))))))............................................................... (-11.52 = -11.82 + 0.30)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:56:26 2011