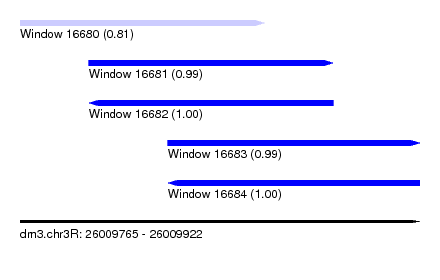
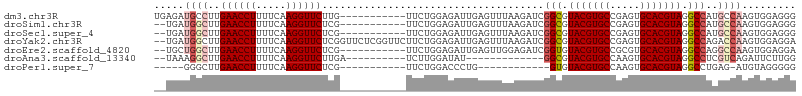
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 26,009,765 – 26,009,922 |
| Length | 157 |
| Max. P | 0.998518 |

| Location | 26,009,765 – 26,009,861 |
|---|---|
| Length | 96 |
| Sequences | 7 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 80.03 |
| Shannon entropy | 0.34442 |
| G+C content | 0.49288 |
| Mean single sequence MFE | -28.27 |
| Consensus MFE | -20.56 |
| Energy contribution | -20.97 |
| Covariance contribution | 0.41 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.805725 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
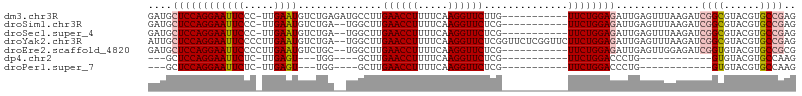
>dm3.chr3R 26009765 96 + 27905053 GAUGCUCCAGGAAUUCCC-UUGAAUGUCUGAGAUGCCUUGAACCUUUUCAAGGUUCUUG-----------UUCUGGAGAUUGAGUUUAAGAUCGGCGUACGUGCCGAG ....((((((((((((..-..))))....((((.((((((((....)))))))))))).-----------)))))))).............((((((....)))))). ( -32.20, z-score = -2.94, R) >droSim1.chr3R 25677228 94 + 27517382 GAUGCUCCAGGAAUUCCC-UUGAAUGUCUGA--UGGCUUGAACCUUUUCAAGGUUCUCG-----------UUCUGGAGAUUGAGUUUAAGAUCGGCGUACGUGCCGAG ....((((((((......-......(((...--.)))..((((((.....))))))...-----------)))))))).............((((((....)))))). ( -28.70, z-score = -1.50, R) >droSec1.super_4 4862517 94 + 6179234 GAUGCUCCAGGAAUUCCC-UUGAAUGUCUGA--UGGCUUGAACCUUUUCAAGGUUCUCG-----------UUCUGGAGAUUGAGUUUAAGAUCGGCGUACGUGCCGAG ....((((((((......-......(((...--.)))..((((((.....))))))...-----------)))))))).............((((((....)))))). ( -28.70, z-score = -1.50, R) >droYak2.chr3R 26943331 106 + 28832112 AUUGCUCCAGGAAUUCCCCUUGAAUGUCUGA--UGGCUUGAACCUUUUCAAGGUUCUCGGUUCUCGGUUCUUCUGGAGAUUGAGUUUAAGAUCGGCGUACGUGCCGAG ....((((((((((((.....))))..((((--..(((.((((((.....))))))..)))..))))...)))))))).............((((((....)))))). ( -33.90, z-score = -1.63, R) >droEre2.scaffold_4820 8481843 95 - 10470090 GAUGCUCCAGGAAUUCCCCUUGAAUGUCUGC--UGGCUUGAACCUUUUCAAGGUUCUCG-----------UUCUGGAGAUUGAGUUGGAGAUCGGUGUACGUGCCGCG (((.((((((....((.((..(((((((...--.)))..((((((.....))))))..)-----------))).)).)).....)))))))))(((......)))... ( -27.00, z-score = -0.33, R) >dp4.chr2 29298012 74 - 30794189 ---GCUCCAGGAAUUCUC-UUGAGU---UGG----GCUUGAACCUUUUCAAGGUUCUCG-----------UUCUGGACCCUG------------GUGUACGUGCCAAG ---((.((((((((((..-..))))---)((----((..((((((.....))))))..)-----------))).....))))------------).)).......... ( -23.70, z-score = -1.54, R) >droPer1.super_7 146153 74 - 4445127 ---GCUCCAGGAAUUCUC-UUGAGU---UGG----GCUUGAACCUUUUCAAGGUUCUCG-----------UUCUGGACCCUG------------GUGUACGUGCCAAG ---((.((((((((((..-..))))---)((----((..((((((.....))))))..)-----------))).....))))------------).)).......... ( -23.70, z-score = -1.54, R) >consensus GAUGCUCCAGGAAUUCCC_UUGAAUGUCUGA__UGGCUUGAACCUUUUCAAGGUUCUCG___________UUCUGGAGAUUGAGUUUAAGAUCGGCGUACGUGCCGAG ....((((((((((((.....))))..............((((((.....))))))..............))))))))..............((((......)))).. (-20.56 = -20.97 + 0.41)
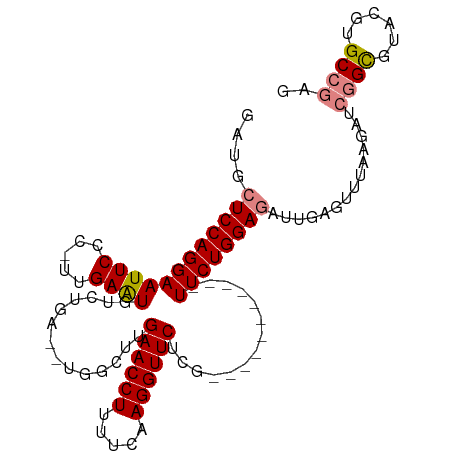
| Location | 26,009,792 – 26,009,888 |
|---|---|
| Length | 96 |
| Sequences | 7 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 77.86 |
| Shannon entropy | 0.40592 |
| G+C content | 0.52288 |
| Mean single sequence MFE | -33.57 |
| Consensus MFE | -24.90 |
| Energy contribution | -25.76 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.985202 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 26009792 96 + 27905053 UGAGAUGCCUUGAACCUUUUCAAGGUUCUUG-----------UUCUGGAGAUUGAGUUUAAGAUCGGCGUACGUGCCGAGUGCACGUAGGCCAUGCCAAGUGGAGGG .((((.((((((((....)))))))))))).-----------...(((.((((........))))(((.(((((((.....))))))).)))...)))......... ( -34.70, z-score = -2.53, R) >droSim1.chr3R 25677255 94 + 27517382 --UGAUGGCUUGAACCUUUUCAAGGUUCUCG-----------UUCUGGAGAUUGAGUUUAAGAUCGGCGUACGUGCCGAGUGCACGUAGGCCAUGCCAAGUGGAGGG --...((((..((((((.....))))))...-----------.......((((........))))(((.(((((((.....))))))).)))..))))......... ( -32.80, z-score = -1.76, R) >droSec1.super_4 4862544 94 + 6179234 --UGAUGGCUUGAACCUUUUCAAGGUUCUCG-----------UUCUGGAGAUUGAGUUUAAGAUCGGCGUACGUGCCGAGUGCACGUAGGCCAUGCCAAGUGGAGGG --...((((..((((((.....))))))...-----------.......((((........))))(((.(((((((.....))))))).)))..))))......... ( -32.80, z-score = -1.76, R) >droYak2.chr3R 26943359 105 + 28832112 --UGAUGGCUUGAACCUUUUCAAGGUUCUCGGUUCUCGGUUCUUCUGGAGAUUGAGUUUAAGAUCGGCGUACGUGCCGAGUGCACGUAGGCCAGACCAAGUGGAGGA --............((((..(..(((((((((((..(((.....)))..))))))).........(((.(((((((.....))))))).))).))))..)..)))). ( -37.80, z-score = -1.71, R) >droEre2.scaffold_4820 8481871 94 - 10470090 --UGCUGGCUUGAACCUUUUCAAGGUUCUCG-----------UUCUGGAGAUUGAGUUGGAGAUCGGUGUACGUGCCGCGUGCACGUAGGCCAGGCCAAGUGGAGGA --.((((((((...((..(((((...((((.-----------.....)))))))))..)).....(((.(((((((.....))))))).)))))))).)))...... ( -35.30, z-score = -1.52, R) >droAna3.scaffold_13340 21043907 82 + 23697760 --UAAAGGCUUGAACCUUUUCAAGGUUCUUGA----------UCUUGGAUAU-------------GGCGUACGUGCCAAGUGCACGUAGGCCUCGUCAGAUUCUUGG --..(((((..((((((.....))))))..).----------)))).(((..-------------(((.(((((((.....))))))).)))..))).......... ( -30.50, z-score = -3.56, R) >droPer1.super_7 146175 78 - 4445127 -----GGGCUUGAACCUUUUCAAGGUUCUCG-----------UUCUGGACCCUG------------GUGUACGUGCCAAGUGCACGUAGGCCUGAG-AUGUAGGGGG -----((((..((((((.....))))))..)-----------)))....(((((------------((.(((((((.....))))))).)))....-....)))).. ( -31.10, z-score = -1.95, R) >consensus __UGAUGGCUUGAACCUUUUCAAGGUUCUCG___________UUCUGGAGAUUGAGUUUAAGAUCGGCGUACGUGCCGAGUGCACGUAGGCCAGGCCAAGUGGAGGG .....((((..((((((.....)))))).....................................(((.(((((((.....))))))).)))..))))......... (-24.90 = -25.76 + 0.86)
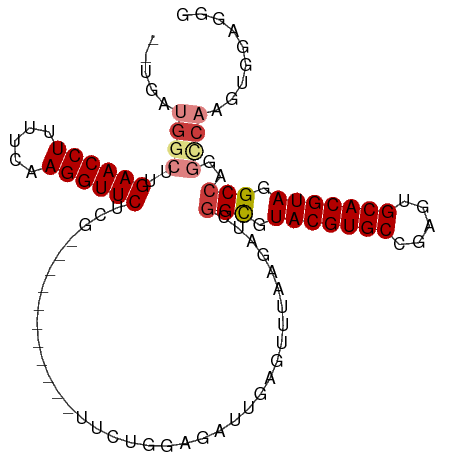
| Location | 26,009,792 – 26,009,888 |
|---|---|
| Length | 96 |
| Sequences | 7 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 77.86 |
| Shannon entropy | 0.40592 |
| G+C content | 0.52288 |
| Mean single sequence MFE | -28.01 |
| Consensus MFE | -21.66 |
| Energy contribution | -21.80 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.39 |
| SVM RNA-class probability | 0.998518 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 26009792 96 - 27905053 CCCUCCACUUGGCAUGGCCUACGUGCACUCGGCACGUACGCCGAUCUUAAACUCAAUCUCCAGAA-----------CAAGAACCUUGAAAAGGUUCAAGGCAUCUCA .......((((...((((.(((((((.....))))))).)))).............((....)).-----------))))..(((((((....)))))))....... ( -27.30, z-score = -2.74, R) >droSim1.chr3R 25677255 94 - 27517382 CCCUCCACUUGGCAUGGCCUACGUGCACUCGGCACGUACGCCGAUCUUAAACUCAAUCUCCAGAA-----------CGAGAACCUUGAAAAGGUUCAAGCCAUCA-- .........((((.((((.(((((((.....))))))).))))......................-----------...((((((.....))))))..))))...-- ( -30.90, z-score = -4.10, R) >droSec1.super_4 4862544 94 - 6179234 CCCUCCACUUGGCAUGGCCUACGUGCACUCGGCACGUACGCCGAUCUUAAACUCAAUCUCCAGAA-----------CGAGAACCUUGAAAAGGUUCAAGCCAUCA-- .........((((.((((.(((((((.....))))))).))))......................-----------...((((((.....))))))..))))...-- ( -30.90, z-score = -4.10, R) >droYak2.chr3R 26943359 105 - 28832112 UCCUCCACUUGGUCUGGCCUACGUGCACUCGGCACGUACGCCGAUCUUAAACUCAAUCUCCAGAAGAACCGAGAACCGAGAACCUUGAAAAGGUUCAAGCCAUCA-- .......(((((((((((.(((((((.....))))))).)))((........))..........)).))))))......((((((.....)))))).........-- ( -31.80, z-score = -2.98, R) >droEre2.scaffold_4820 8481871 94 + 10470090 UCCUCCACUUGGCCUGGCCUACGUGCACGCGGCACGUACACCGAUCUCCAACUCAAUCUCCAGAA-----------CGAGAACCUUGAAAAGGUUCAAGCCAGCA-- ........(((((((((..(((((((.....)))))))....((........)).....))))..-----------...((((((.....))))))..)))))..-- ( -26.80, z-score = -2.08, R) >droAna3.scaffold_13340 21043907 82 - 23697760 CCAAGAAUCUGACGAGGCCUACGUGCACUUGGCACGUACGCC-------------AUAUCCAAGA----------UCAAGAACCUUGAAAAGGUUCAAGCCUUUA-- ....((.(((...(((((.(((((((.....))))))).)))-------------...))..)))----------))..((((((.....)))))).........-- ( -25.40, z-score = -3.44, R) >droPer1.super_7 146175 78 + 4445127 CCCCCUACAU-CUCAGGCCUACGUGCACUUGGCACGUACAC------------CAGGGUCCAGAA-----------CGAGAACCUUGAAAAGGUUCAAGCCC----- ..((((....-....((..(((((((.....)))))))..)------------))))).......-----------...((((((.....))))))......----- ( -23.00, z-score = -1.57, R) >consensus CCCUCCACUUGGCAUGGCCUACGUGCACUCGGCACGUACGCCGAUCUUAAACUCAAUCUCCAGAA___________CGAGAACCUUGAAAAGGUUCAAGCCAUCA__ .............(((((.(((((((.....))))))).........................................((((((.....))))))..))))).... (-21.66 = -21.80 + 0.14)

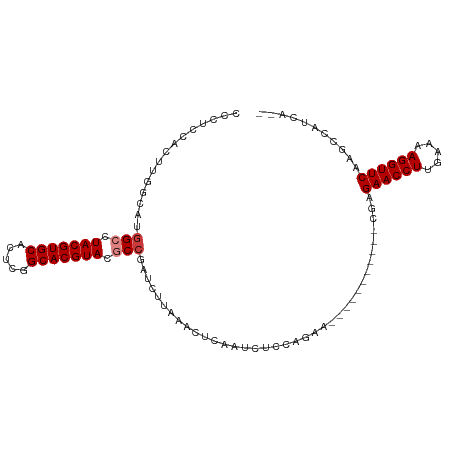
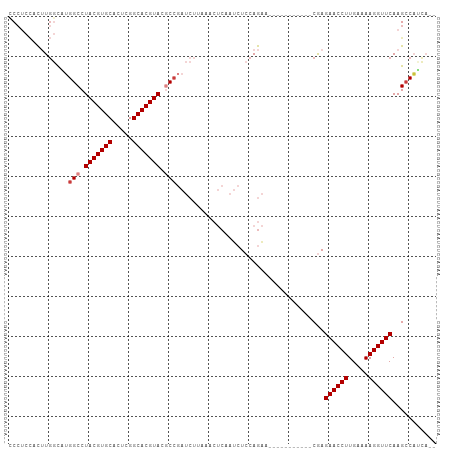
| Location | 26,009,823 – 26,009,922 |
|---|---|
| Length | 99 |
| Sequences | 8 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 62.94 |
| Shannon entropy | 0.64806 |
| G+C content | 0.54893 |
| Mean single sequence MFE | -32.19 |
| Consensus MFE | -16.37 |
| Energy contribution | -16.39 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.45 |
| SVM RNA-class probability | 0.991069 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 26009823 99 + 27905053 --UUCUGGAGAUUGAGUUUAAGAUCGGCGUACGUGCCGAGUGCACGUAGGCCAUGCC-----------AAGUGGAGGGAGUCAAGGGG------CAGGUGGGGAAGUUAGUUAAUUGA --.......((((((.(((...((((((.(((((((.....))))))).))).((((-----------...(((......)))...))------)))))...))).))))))...... ( -32.50, z-score = -2.89, R) >droSim1.chr3R 25677284 99 + 27517382 --UUCUGGAGAUUGAGUUUAAGAUCGGCGUACGUGCCGAGUGCACGUAGGCCAUGCC-----------AAGUGGAGGGUGUCAAGGGG------CAGGUGGGGAAGUUAGUUAAUUGA --.......((((((.(((...((((((.(((((((.....))))))).))).((((-----------...(((......)))...))------)))))...))).))))))...... ( -32.50, z-score = -2.69, R) >droSec1.super_4 4862573 99 + 6179234 --UUCUGGAGAUUGAGUUUAAGAUCGGCGUACGUGCCGAGUGCACGUAGGCCAUGCC-----------AAGUGGAGGGAGUCGAGGGG------CAGGUGGGGAAGUUAGUUAAUUGA --.......((((((.(((...((((((.(((((((.....))))))).))).((((-----------...(((......)))...))------)))))...))).))))))...... ( -31.80, z-score = -2.50, R) >droYak2.chr3R 26943397 107 + 28832112 UCUUCUGGAGAUUGAGUUUAAGAUCGGCGUACGUGCCGAGUGCACGUAGGCCAGACC-----------AAGUGGAGGAUCCCAGGGGGGGAUCUCAGGUGGGGAAGUUAGUUAAUUGA .(((((...((((........))))(((.(((((((.....))))))).)))...((-----------(..((..(((((((.....)))))))))..))))))))............ ( -39.80, z-score = -2.72, R) >droEre2.scaffold_4820 8481900 99 - 10470090 --UUCUGGAGAUUGAGUUGGAGAUCGGUGUACGUGCCGCGUGCACGUAGGCCAGGCC-----------AAGUGGAGGAUCUUGGGGGC------UAGGCGGGGAAGUUAGUUAAUUGA --((((.(.....(..((.(((((((((.(((((((.....))))))).)))...((-----------....))..)))))).))..)------....).)))).............. ( -30.20, z-score = -0.71, R) >droAna3.scaffold_13340 21043937 81 + 23697760 ---------------UCUUGGAUAUGGCGUACGUGCCAAGUGCACGUAGGCCUCGUC-----------AGAUUCUUGGAGCUCAGGGG------CAAGUGGUGGAGC-AAUCAA---- ---------------.(((((((..(((.(((((((.....))))))).)))..)))-----------...((((((.....))))))------))))((((.....-.)))).---- ( -27.70, z-score = -1.21, R) >dp4.chr2 29298060 99 - 30794189 --------------UUCUGGACCCUGGUGUACGUGCCAAGUGCACGUAGGCACUGCCAGAAGGUCCUGAGAUGUAGGGGGAAGAGAAGGG-CCGCUGGUGCAGGUGGAAAUCGA---- --------------(((((...((((((.(((((((.....))))))).))...(((((..((((((...................))))-)).))))).))))))))).....---- ( -35.11, z-score = -0.74, R) >droPer1.super_7 146201 85 - 4445127 --------------UUCUGGACCCUGGUGUACGUGCCAAGUGCACGUAGGC--------------CUGAGAUGUAGGGGGAAGAGAAGGG-GCGCUGGUGCAGGUGGAAAUCGA---- --------------((((...(((((((.(((((((.....))))))).))--------------)........))))...)))).....-(((....))).(((....)))..---- ( -27.90, z-score = -0.71, R) >consensus __UUCUGGAGAUUGAGUUUAAGAUCGGCGUACGUGCCGAGUGCACGUAGGCCAUGCC___________AAGUGGAGGGAGUCGAGGGG______CAGGUGGGGAAGUUAGUUAAUUGA .........................(((.(((((((.....))))))).))).................................................................. (-16.37 = -16.39 + 0.02)

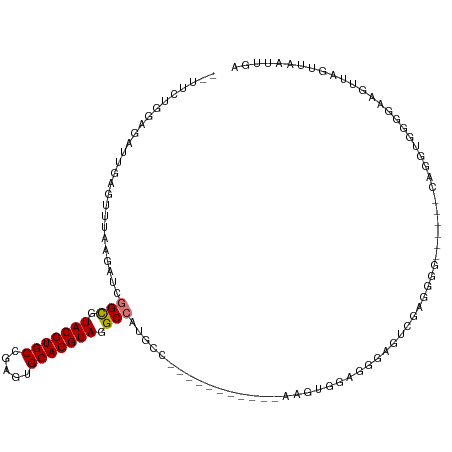
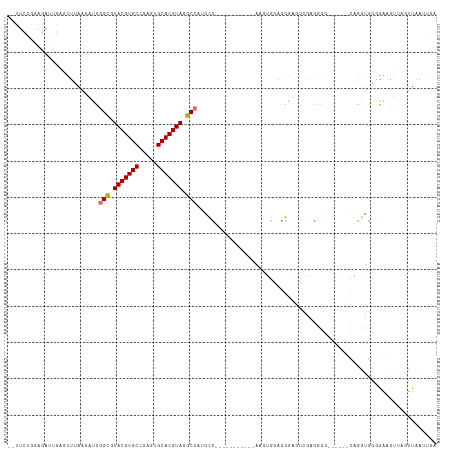
| Location | 26,009,823 – 26,009,922 |
|---|---|
| Length | 99 |
| Sequences | 8 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 62.94 |
| Shannon entropy | 0.64806 |
| G+C content | 0.54893 |
| Mean single sequence MFE | -22.38 |
| Consensus MFE | -10.95 |
| Energy contribution | -11.20 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.24 |
| SVM RNA-class probability | 0.998017 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 26009823 99 - 27905053 UCAAUUAACUAACUUCCCCACCUG------CCCCUUGACUCCCUCCACUU-----------GGCAUGGCCUACGUGCACUCGGCACGUACGCCGAUCUUAAACUCAAUCUCCAGAA-- ......................((------((...((........))...-----------))))((((.(((((((.....))))))).))))......................-- ( -21.30, z-score = -3.47, R) >droSim1.chr3R 25677284 99 - 27517382 UCAAUUAACUAACUUCCCCACCUG------CCCCUUGACACCCUCCACUU-----------GGCAUGGCCUACGUGCACUCGGCACGUACGCCGAUCUUAAACUCAAUCUCCAGAA-- ......................((------((...((........))...-----------))))((((.(((((((.....))))))).))))......................-- ( -21.30, z-score = -3.41, R) >droSec1.super_4 4862573 99 - 6179234 UCAAUUAACUAACUUCCCCACCUG------CCCCUCGACUCCCUCCACUU-----------GGCAUGGCCUACGUGCACUCGGCACGUACGCCGAUCUUAAACUCAAUCUCCAGAA-- ......................((------((....((.....)).....-----------))))((((.(((((((.....))))))).))))......................-- ( -20.10, z-score = -2.98, R) >droYak2.chr3R 26943397 107 - 28832112 UCAAUUAACUAACUUCCCCACCUGAGAUCCCCCCCUGGGAUCCUCCACUU-----------GGUCUGGCCUACGUGCACUCGGCACGUACGCCGAUCUUAAACUCAAUCUCCAGAAGA ............((((.......((((((((.....))))).))).....-----------((((.(((.(((((((.....))))))).)))))))................)))). ( -32.70, z-score = -4.43, R) >droEre2.scaffold_4820 8481900 99 + 10470090 UCAAUUAACUAACUUCCCCGCCUA------GCCCCCAAGAUCCUCCACUU-----------GGCCUGGCCUACGUGCACGCGGCACGUACACCGAUCUCCAACUCAAUCUCCAGAA-- ...................(((.(------(...(((((........)))-----------)).))))).(((((((.....)))))))...........................-- ( -18.90, z-score = -1.62, R) >droAna3.scaffold_13340 21043937 81 - 23697760 ----UUGAUU-GCUCCACCACUUG------CCCCUGAGCUCCAAGAAUCU-----------GACGAGGCCUACGUGCACUUGGCACGUACGCCAUAUCCAAGA--------------- ----(((...-((((.........------.....))))..)))......-----------.....(((.(((((((.....))))))).)))..........--------------- ( -20.44, z-score = -1.59, R) >dp4.chr2 29298060 99 + 30794189 ----UCGAUUUCCACCUGCACCAGCGG-CCCUUCUCUUCCCCCUACAUCUCAGGACCUUCUGGCAGUGCCUACGUGCACUUGGCACGUACACCAGGGUCCAGAA-------------- ----...........((((....))))-........................(((((((..(((...)))(((((((.....)))))))....)))))))....-------------- ( -26.30, z-score = -0.55, R) >droPer1.super_7 146201 85 + 4445127 ----UCGAUUUCCACCUGCACCAGCGC-CCCUUCUCUUCCCCCUACAUCUCAG--------------GCCUACGUGCACUUGGCACGUACACCAGGGUCCAGAA-------------- ----...........(((.(((...((-(.......................)--------------)).(((((((.....)))))))......))).)))..-------------- ( -18.00, z-score = -0.51, R) >consensus UCAAUUAACUAACUUCCCCACCUG______CCCCUCGACACCCUCCACUU___________GGCAUGGCCUACGUGCACUCGGCACGUACGCCGAUCUCAAACUCAAUCUCCAGAA__ ..................................................................((..(((((((.....)))))))..))......................... (-10.95 = -11.20 + 0.25)
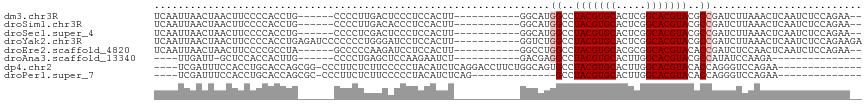

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:56:24 2011