| Sequence ID | dm3.chr3R |
|---|---|
| Location | 25,967,950 – 25,968,053 |
| Length | 103 |
| Max. P | 0.941191 |

| Location | 25,967,950 – 25,968,053 |
|---|---|
| Length | 103 |
| Sequences | 7 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 61.52 |
| Shannon entropy | 0.71786 |
| G+C content | 0.44979 |
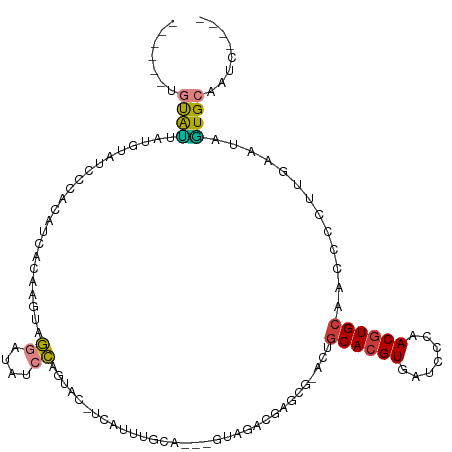
| Mean single sequence MFE | -27.00 |
| Consensus MFE | -8.78 |
| Energy contribution | -8.70 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.50 |
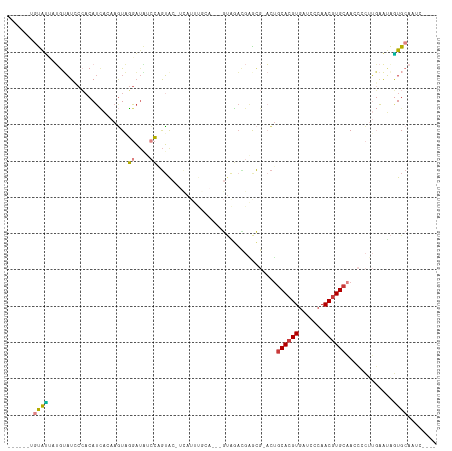
| Mean z-score | -1.93 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.941191 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
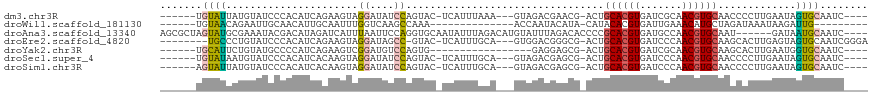
>dm3.chr3R 25967950 103 - 27905053 ------UGUAUUAUGUAUCCCACAUCAGAAGUAGGAUAUCCAGUAC-UCAUUUAAA---GUAGACGAACG-ACUGCACGUGAUCGCAACGUGCAACCCCUUGAAUAGUGCAAUC---- ------........((((((.((.......)).))))))...((((-(.((((((.---((......)).-..(((((((.......))))))).....)))))))))))....---- ( -23.70, z-score = -1.64, R) >droWil1.scaffold_181130 9422982 88 - 16660200 ------UGUAACAGAAUUGCAACAUUGCAAUUUGGUCAAGCCAAA--------------ACCAAUACAUA-CAUACACGUGAUUGAAACAUGCUAGAUAAAUAAGAUUG--------- ------.......((((((((....)))))))).(((.(((....--------------..((((.(((.-.......)))))))......))).)))...........--------- ( -14.00, z-score = -0.88, R) >droAna3.scaffold_13340 20957838 108 - 23697760 AGCGCUAGUAUGCGAAAUACGACAUAGAUCAUUUAAUUCCAGGUGCAAUAUUUAGACAUGUAUUUAGACACCCCGCACGUGAUGCCAACGUGCAAU------GAUAAUGCAAUC---- .(((....((((((.....)).)))).((((((........((((((((((........)))))..).))))..((((((.......)))))))))------)))..)))....---- ( -23.00, z-score = -0.69, R) >droEre2.scaffold_4820 8437838 104 + 10470090 --------UGCCCUGUAUCCCACAUCAGAAGUAGGAUAGCC-GUAC-UCAUUUGCA---GUGGACGGGCG-ACUGCACGUGAUCCCAACGUGCAAGCACUUGAGUAGUGCAAUCGGGA --------..(((..(((((.((.......)).)))))((.-.(((-(((..((((---((.(.....).-)))((((((.......))))))..)))..))))))..))....))). ( -35.90, z-score = -1.42, R) >droYak2.chr3R 26899835 90 - 28832112 ------UGCAUUCUGUAUGCCCCAUCAGAAGUCGGAUGUCCAGUG-----------------GAGGAGCG-ACUGCACGUGAUCGCAACGUGCAAGCACUUGAAUGGUGCAAUC---- ------....(((((.(((...))))))))((((..(.(((...)-----------------)).)..))-))(((((((.......))))))).(((((.....)))))....---- ( -28.40, z-score = -0.40, R) >droSec1.super_4 4820753 103 - 6179234 ------UGUAUAAUGUAUCCCACAUCACAAGUAGGAUAUCCAGUAC-UCAUUUGCA---GUAGACGAGCG-ACUGCACGUGAUCCCAACGUGCAACCCCUUGAAUAGUGCAAUC---- ------.((((...((((((.((.......)).))))))...))))-....(((((---.((..((((.(-..(((((((.......)))))))..).))))..)).)))))..---- ( -31.00, z-score = -3.83, R) >droSim1.chr3R 25635396 103 - 27517382 ------AGUAUUAUGUAUCCCACAUCACAAGUAGGAUAUCCAGUAC-UCAUUUGCA---GUAGACGAGCG-ACUGCACGUGAUCCCAACGUGCAACCCCUUGAAUAGUGCAAUC---- ------((((((..((((((.((.......)).))))))..)))))-)...(((((---.((..((((.(-..(((((((.......)))))))..).))))..)).)))))..---- ( -33.00, z-score = -4.63, R) >consensus ______UGUAUUAUGUAUCCCACAUCACAAGUAGGAUAUCCAGUAC_UCAUUUGCA___GUAGACGAGCG_ACUGCACGUGAUCCCAACGUGCAACCCCUUGAAUAGUGCAAUC____ .......((((......................((....)).................................((((((.......)))))).............))))........ ( -8.78 = -8.70 + -0.08)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:56:20 2011