| Sequence ID | dm3.chr2L |
|---|---|
| Location | 9,195,211 – 9,195,330 |
| Length | 119 |
| Max. P | 0.925693 |

| Location | 9,195,211 – 9,195,330 |
|---|---|
| Length | 119 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.98 |
| Shannon entropy | 0.50859 |
| G+C content | 0.45392 |
| Mean single sequence MFE | -34.06 |
| Consensus MFE | -22.73 |
| Energy contribution | -22.63 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.925693 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 9195211 119 + 23011544 UUUUUUGCUCGCCCUAAUUGUGCUGGGCGUGGCAGGCAAAUCAAAUUUGUAAACAGAUUUCGUUGCCACGGAAAGGGGGCAA-AUAUAAAGACCCUCACUGGGUUGUCUUUGCUAAAUUU .....(((((((((..........))))).))))(((((...(((((((....)))))))..)))))..........(((((-(......(((((.....)))))...))))))...... ( -38.20, z-score = -2.03, R) >droSim1.chr2L 8976578 118 + 22036055 UUUUUUGCUCGCCCUAAUUGUGCUGGGCGUGGCAGGCAAAUCAAAUUUGUAAACAGAUUUCGUUGCCACGGAAGGGGGCAAU-AUAUAAAGACCCUCACUGGGUUG-UCUUGCUAAAUUU .....(((((((((..........))))).))))(((((..((((((((....))))))).((((((.(....)..))))))-.......(((((.....))))))-..)))))...... ( -39.30, z-score = -2.38, R) >droSec1.super_3 4660092 118 + 7220098 UUUUUUGCUCGCCCUAAUUGUGCUGGGCGUGGCAGGCAAAUCAAAUUUGUAAACAGAUUUCGUUGCCACGGAAGGGGGCAAU-AUAUAAAGACCCUCACUGGGUUG-UCUUGCUAAAUUU .....(((((((((..........))))).))))(((((..((((((((....))))))).((((((.(....)..))))))-.......(((((.....))))))-..)))))...... ( -39.30, z-score = -2.38, R) >droYak2.chr2L 11864449 117 + 22324452 UUUUUUGCUCGCCCUAAUUGCGCUGGGCGUGGCAGGCAAAUCAAAUUUGUAAACAGAUUUCGUUGCCACGGAAAGGGGG-GG-AUAUAAAGACCCUCUCUGGGUUG-CUUUGCUAAAUUU .....(((((((((..........))))).))))((((((..(((((((....))))))).((.(((.((((..(((((-..-.........)))))))))))).)-)))))))...... ( -38.50, z-score = -1.63, R) >droEre2.scaffold_4929 9803898 119 + 26641161 UUUUUUGCUCGCCCUAAUUGCGCUGGGCGUGGCAGGCAAAUCAAAUUUGUAAACAGAUUUCGUUGCCACGGAAGGGGGCAAGUGUAUAAAGAGGCUCUCUGGGUCG-CCUUGCUAAAUUU .....(((((((((..........))))).))))(((((...(((((((....)))))))..))))).(....)..((((((.((((..((((...))))..)).)-)))))))...... ( -39.30, z-score = -1.17, R) >droAna3.scaffold_12943 542748 107 + 5039921 -UUUUUGCUCGCCCUAAUUGCGCUGGGCGUGGCAGGCAAAUCAAAUUUGUAAACAGAUUUCGUUGCCACCGGGUGGGGG-------UAAACACUCUC----GGUCG-UGUUCUUAAAUUU -....(((((((((..........))))).))))(((((...(((((((....)))))))..)))))(((((.((((((-------(....))))))----).)))-.)).......... ( -37.30, z-score = -2.84, R) >dp4.chr4_group5 1510802 96 + 2436548 --UUUUUUGCGCCCUAAUUGCACUGGGCGUGGCAGGCAAAUCAAAUUGGCAAACAGAUUUUGUUGUUUUUCGAGGACUGUUGAGGGGAUUCACGUUUC---------------------- --....(..(((((..........)))))..)..(.(((......))).).(((((.(((((........))))).))))).................---------------------- ( -20.30, z-score = 1.12, R) >droPer1.super_5 5919334 96 + 6813705 --UUUUUUGCGCCCUAAUUGCACUGGGCGUGGCAGGCAAAUCAAAUUGGCAAACAGAUUUUGUUGUUUUUCGAGGACUGUUGAGGGGAUUCACGUUUC---------------------- --....(..(((((..........)))))..)..(.(((......))).).(((((.(((((........))))).))))).................---------------------- ( -20.30, z-score = 1.12, R) >consensus UUUUUUGCUCGCCCUAAUUGCGCUGGGCGUGGCAGGCAAAUCAAAUUUGUAAACAGAUUUCGUUGCCACGGAAGGGGGGAAG_AUAUAAAGACCCUCACUGGGUUG_CCUUGCUAAAUUU .....(((((((((..........))))).))))(((((...(((((((....)))))))..))))).......(((((.............)))))....................... (-22.73 = -22.63 + -0.09)

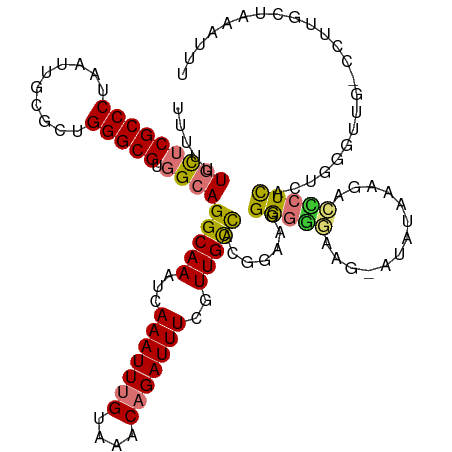
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:27:42 2011