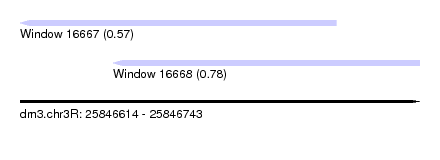
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 25,846,614 – 25,846,743 |
| Length | 129 |
| Max. P | 0.779514 |

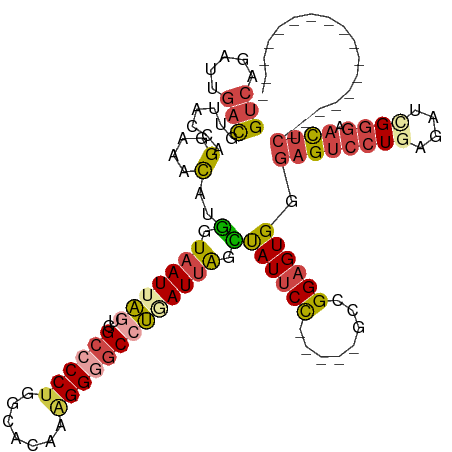
| Location | 25,846,614 – 25,846,716 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 124 |
| Reading direction | reverse |
| Mean pairwise identity | 74.14 |
| Shannon entropy | 0.44374 |
| G+C content | 0.51829 |
| Mean single sequence MFE | -34.58 |
| Consensus MFE | -19.39 |
| Energy contribution | -19.12 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.573828 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 25846614 102 - 27905053 GUCAGAUUGACGUUACCAGGAAACAUGGUAAUUAGUGGCCCCUGGCACAAAGGGGCGUGAUUAGCUAUUCC-----GCCGGAGUGAGAGUCCUGAGAUCGGGAACUC----------------- (((.....)))((((((((....).))))))).....((((((.......)))))).........((((((-----...)))))).((((((((....)))).))))----------------- ( -36.50, z-score = -2.14, R) >droSim1.chr3R_random 1249923 102 - 1307089 GUCAGAUUGACGUUACCAGGAAACAUGGUAAUUAGUGGCCCCUGGCACAAAGGGGCCUGAUUAGCUAUUCC-----GCCGGAGUGGGAGUCCUGAGAUCGGGAACUC----------------- (((.....)))((((((((....).)))))))....(((((((.......))))))).......(((((((-----...)))))))((((((((....)))).))))----------------- ( -42.30, z-score = -3.15, R) >droSec1.super_4 4692460 102 - 6179234 GUCAGAUUGACGUUACCAGGAAACAUGGUAAUUAGUGGCCCCUGGCACAAAGGGGCCUGAUUAGCUAUUCC-----GCCGGAGUGGGAGUCCUGAGAUCGGGAACUC----------------- (((.....)))((((((((....).)))))))....(((((((.......))))))).......(((((((-----...)))))))((((((((....)))).))))----------------- ( -42.30, z-score = -3.15, R) >droYak2.chr3R 26769386 107 - 28832112 GUCAGAUUGACGUUGCCAGGAAACAUUCUAAUUAGUUGCCCCCGGCACAACGGGGCCUCAUUACGCAUUCCUCCGGAGUGGAGUGGGAGUCCUAGGAUCGGGAACUC----------------- (((.....)))(((.((.(....)(((((.....((.(((...)))))...(((((((((((.(.((((((...))))))))))))).)))))))))).)).)))..----------------- ( -34.90, z-score = -0.34, R) >droEre2.scaffold_4820 8308891 102 + 10470090 GUCAGAUUGACGUUACCAGGAAACAUUGUAAUUAGCUGCCCCUGGCACAAAGGUGCCUCAUUACGUAUUCCU-----GCGGAGUGAGAGUCCUAGGAUCGGGAACCC----------------- (((.....)))(((((..(....)...))))).......(((.(((((....)))))...........((((-----(.(((.(....)))))))))..))).....----------------- ( -27.00, z-score = -0.00, R) >droAna3.scaffold_13340 20855071 118 - 23697760 GAUAGACUCAUGUUUUCAGGAAGUAUUAUAAUUUGUUG--CCUGGCACUGGGGAACGAAAUCGGAUAUCCUU-UAGGGAGGAGUGGGAG---UGGGAGUGAGAGUUAAUUGUAAUCAAGCAGGC ....(((((.(..(..((.....((((....((((((.--(((......))).))))))....))))(((((-....))))).......---))..)..).)))))..((((......)))).. ( -24.50, z-score = 0.52, R) >consensus GUCAGAUUGACGUUACCAGGAAACAUGGUAAUUAGUGGCCCCUGGCACAAAGGGGCCUGAUUAGCUAUUCC_____GCCGGAGUGGGAGUCCUGAGAUCGGGAACUC_________________ (((.....)))(((.((.(....)..(.(((((((..((((((.......))))))))))))).)((((((........))))))..............)).)))................... (-19.39 = -19.12 + -0.27)

| Location | 25,846,644 – 25,846,743 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.22 |
| Shannon entropy | 0.42640 |
| G+C content | 0.47526 |
| Mean single sequence MFE | -29.37 |
| Consensus MFE | -17.10 |
| Energy contribution | -17.30 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.779514 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 25846644 99 - 27905053 ----------GUUCAUUGAUUUUCCG---CACUUAAGUGCGUCAGAUUGACGUUACCAGGAAACAUGGUAAUUAGUGGCCCCUGGCACAAAGGGGCGUGAUUAGCUAUUCCG-------- ----------..((((..((((..((---(((....)))))..))))..).((((((((....).))))))).....((((((.......)))))).)))............-------- ( -33.40, z-score = -2.79, R) >droSim1.chr3R_random 1249953 99 - 1307089 ----------GUUCAUUGAUUUUCCG---CACUUAAGUGCGUCAGAUUGACGUUACCAGGAAACAUGGUAAUUAGUGGCCCCUGGCACAAAGGGGCCUGAUUAGCUAUUCCG-------- ----------.....(..((((..((---(((....)))))..))))..).((((((((....).)))))))(((((((((((.......)))))))......)))).....-------- ( -36.20, z-score = -3.71, R) >droSec1.super_4 4692490 99 - 6179234 ----------GUUCAUUGAUUUUCCG---CACUUAAGUGCGUCAGAUUGACGUUACCAGGAAACAUGGUAAUUAGUGGCCCCUGGCACAAAGGGGCCUGAUUAGCUAUUCCG-------- ----------.....(..((((..((---(((....)))))..))))..).((((((((....).)))))))(((((((((((.......)))))))......)))).....-------- ( -36.20, z-score = -3.71, R) >droYak2.chr3R 26769416 104 - 28832112 ----------GCCCAUUGAUUUUCCC---CACUCAAGUGCGUCAGAUUGACGUUGCCAGGAAACAUUCUAAUUAGUUGCCCCCGGCACAACGGGGCCUCAUUACGCAUUCCUCCGGA--- ----------((....(((....(((---(........(((((.....)))))((((.((.(((..........)))...)).))))....))))..)))....))...........--- ( -24.10, z-score = 0.17, R) >droEre2.scaffold_4820 8308921 99 + 10470090 ----------GCUCAUUGAUUUUCCC---CACUCGAGUGCGUCAGAUUGACGUUACCAGGAAACAUUGUAAUUAGCUGCCCCUGGCACAAAGGUGCCUCAUUACGUAUUCCU-------- ----------................---.....(((((((((((.(((..(((((..(....)...))))))))))).....(((((....))))).....))))))))..-------- ( -21.30, z-score = -0.44, R) >droAna3.scaffold_13340 20855111 118 - 23697760 UGAGCAGGAGGUUCAUUGAUUUUGUUUAGCAUUUGAGGGGGAUAGACUCAUGUUUUCAGGAAGUAUUAUAAUUUGUUG--CCUGGCACUGGGGAACGAAAUCGGAUAUCCUUUAGGGAGG (((((.....)))))((((((((((((..((..((((.........))))((((..(((.((((......)))).)))--...)))).))..))))))))))))...(((.....))).. ( -25.00, z-score = 0.61, R) >consensus __________GUUCAUUGAUUUUCCG___CACUUAAGUGCGUCAGAUUGACGUUACCAGGAAACAUGGUAAUUAGUGGCCCCUGGCACAAAGGGGCCUGAUUAGCUAUUCCG________ ....................................(((((((.....)))).)))..((((....(.(((((((..((((((.......))))))))))))).)..))))......... (-17.10 = -17.30 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:56:11 2011