
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 25,805,424 – 25,805,539 |
| Length | 115 |
| Max. P | 0.998417 |

| Location | 25,805,424 – 25,805,539 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 53.98 |
| Shannon entropy | 0.64060 |
| G+C content | 0.31620 |
| Mean single sequence MFE | -22.78 |
| Consensus MFE | -8.00 |
| Energy contribution | -8.67 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.21 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.35 |
| SVM RNA-class probability | 0.998417 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
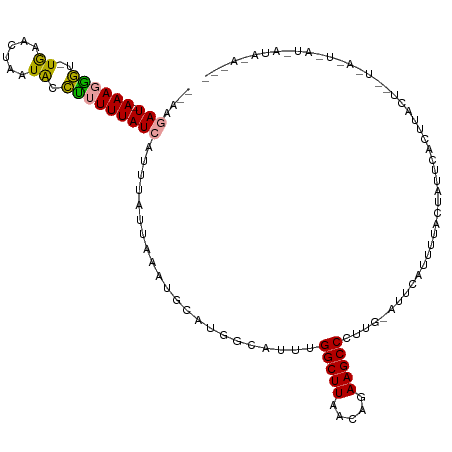
>dm3.chr3R 25805424 115 + 27905053 CCAAGAU-AAAGAGU-UGGACUAAUACCUUUUUAUCAUUUAUUAAGUGCAUGGCAUUUGGCUUUACAGAAGCCCUUGGAUUCGUCCAACCAUUCAGUAACUUUUUAUUAAUCAUAUA--- ....(((-(((((((-(((((.(((.((...............((((((...))))))((((((...))))))...))))).)))).((......))))))))))))).........--- ( -24.90, z-score = -2.22, R) >droYak2.chr3R 8162257 105 - 28832112 --AUGAUCAAAUGAU-UGAAUUAAUACUUUUUUAUCGACCUUUAAUUG----------GGCUUCUCAGAAGCCCUUG-AUGCUUUUUACUAUUCACUUAUUAAUCACUUAU-AUACACAC --.((((..(((((.-(((((...........((((((.........(----------((((((...))))))))))-))).........))))).))))).)))).....-........ ( -18.05, z-score = -3.45, R) >droSim1.chr3R 25473655 96 + 27517382 --CGAAUAAAAGGGCAUAAACUAAUGCCCUUUUAUCAUUUAUUAAAAGCAUGGCAUUUGGCUUAACAGAAGCCCUC--ACUAACUUUAUUAGUUAUAUAC-------------------- --.((.(((((((((((......)))))))))))))...............(((.((((......)))).)))...--..(((((.....))))).....-------------------- ( -25.40, z-score = -3.95, R) >consensus __AAGAU_AAAGGGU_UGAACUAAUACCUUUUUAUCAUUUAUUAAAUGCAUGGCAUUUGGCUUAACAGAAGCCCUUG_AUUCAUUUUACUAUUCACUUACU__U_A_U_AU_AUA_A___ ....(((.(((((((.(......).))))))).)))......................(((((.....)))))............................................... ( -8.00 = -8.67 + 0.67)

| Location | 25,805,424 – 25,805,539 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 53.98 |
| Shannon entropy | 0.64060 |
| G+C content | 0.31620 |
| Mean single sequence MFE | -23.67 |
| Consensus MFE | -7.33 |
| Energy contribution | -6.67 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.939215 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 25805424 115 - 27905053 ---UAUAUGAUUAAUAAAAAGUUACUGAAUGGUUGGACGAAUCCAAGGGCUUCUGUAAAGCCAAAUGCCAUGCACUUAAUAAAUGAUAAAAAGGUAUUAGUCCA-ACUCUUU-AUCUUGG ---......................((((.((((((((.........(((((.....))))).((((((.......................)))))).)))))-))).)))-)...... ( -23.60, z-score = -1.30, R) >droYak2.chr3R 8162257 105 + 28832112 GUGUGUAU-AUAAGUGAUUAAUAAGUGAAUAGUAAAAAGCAU-CAAGGGCUUCUGAGAAGCC----------CAAUUAAAGGUCGAUAAAAAAGUAUUAAUUCA-AUCAUUUGAUCAU-- ........-....((((((((....(((((.((.....))..-...(((((((...))))))----------)..........................)))))-.....))))))))-- ( -20.80, z-score = -2.67, R) >droSim1.chr3R 25473655 96 - 27517382 --------------------GUAUAUAACUAAUAAAGUUAGU--GAGGGCUUCUGUUAAGCCAAAUGCCAUGCUUUUAAUAAAUGAUAAAAGGGCAUUAGUUUAUGCCCUUUUAUUCG-- --------------------((((...((((((...))))))--...(((((.....))))).......))))...........(((((((((((((......)))))))))))))..-- ( -26.60, z-score = -3.09, R) >consensus ___U_UAU_AU_A_U_A__AGUAAAUGAAUAGUAAAAAGAAU_CAAGGGCUUCUGUAAAGCCAAAUGCCAUGCAAUUAAUAAAUGAUAAAAAGGUAUUAGUUCA_ACCCUUU_AUCAU__ ...............................................(((((.....)))))......................(((.(((.(((..........))).))).))).... ( -7.33 = -6.67 + -0.66)



Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:56:04 2011