
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 25,794,636 – 25,794,757 |
| Length | 121 |
| Max. P | 0.586714 |

| Location | 25,794,636 – 25,794,757 |
|---|---|
| Length | 121 |
| Sequences | 7 |
| Columns | 143 |
| Reading direction | forward |
| Mean pairwise identity | 61.43 |
| Shannon entropy | 0.68877 |
| G+C content | 0.45285 |
| Mean single sequence MFE | -31.83 |
| Consensus MFE | -11.27 |
| Energy contribution | -11.94 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.586714 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
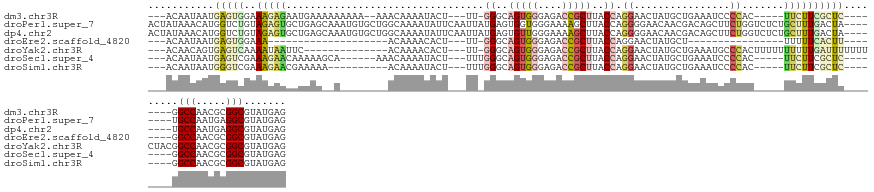
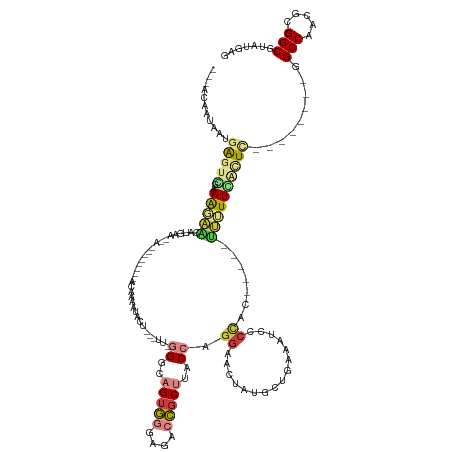
>dm3.chr3R 25794636 121 + 27905053 ---ACAAUAAUGAGUGGAAAGAGAAUGAAAAAAAAA--AAACAAAAUACU---UU-GGGCAGUGGGAGACCGCUUACCAGGAACUAUGCUGAAAUCCCCAC-----UUCUUCGCUC--------GGCCAACGCGGCGUAUGAG ---.......(((((((((((...............--............---.(-((..(((((....)))))..)))(((............)))...)-----)).)))))))--------)(((.....)))....... ( -28.80, z-score = -1.07, R) >droPer1.super_7 3691060 135 + 4445127 ACUAUAAACAUGGUCUGUAGAGUGCUGAGCAAAUGUGCUGGCAAAAUAUUCAAUUAUGAGUGGUGGGAAAAGCUUACCAGGGGAACAACGACAGCUUCUGGUCUCUGCUUUGACUA--------UGCCAAUGAGGCGUAUGAG (((((...((((((..(((...((((.((((....))))))))...)))...)))))).)))))((..(((((..((((((((...........))))))))....)))))..))(--------((((.....)))))..... ( -37.20, z-score = -0.65, R) >dp4.chr2 905084 135 + 30794189 ACUAUAAACAUGGUCUGUAGAGUGCUGAGCAAAUGUGCUGGCAAAAUAUUCAAUUAUGAGUGUUGGGAAAAGCUUACCAGGGGAACAACGACAGCUUCUGGUCUCUGCUUUGACUA--------UGCCAAUGAGGCGUAUGAG .......((((((((.((((((((((.((((....)))))))).((((((((....))))))))...........((((((((...........))))))))))))))...)))))--------)(((.....)))))..... ( -39.10, z-score = -1.33, R) >droEre2.scaffold_4820 8256983 92 - 10470090 ---ACAAUAAUGAGUGGAAA--------------------ACAAAACACU---UU-GGGCAGUGGGAGACCGCUUACCAGGAACUAUGCU----------------UUUUUCACUU--------GGCCAACGCGGCGUAUGAG ---........(((((((((--------------------(........(---((-((..(((((....)))))..))))).........----------------))))))))))--------.(((.....)))....... ( -27.43, z-score = -2.46, R) >droYak2.chr3R 8150944 122 - 28832112 ---ACAACAGUGAGUCAAAAUAAUUC--------------ACAAAACACU---UU-GGGCAGUGGGAGACCGCUUACCAGGAACUAUGCUGAAAUGCCCACUUUUUUUUUUGAUUUUUUUCUACGGCCAACGCGGCGUAUGAG ---.(((..((((((.......))))--------------))........---.(-(((((((((....))))....(((........)))...)))))).........))).......((((((.((.....)))))).)). ( -31.20, z-score = -1.61, R) >droSec1.super_4 4640246 118 + 6179234 ---ACAAUAAUGAGUCGAAAGAACAAAAAGCA------AAACAAAAUACU---UUUGGGCAGUGGGAGACCGCUUACCAGGAACUAUGCUGAAAUCCCCAC-----UUCUUCGCUC--------GGCCAACGCGGCGUAUGAG ---.......((((.((((.........((((------...........(---(((((..(((((....)))))..))))))....))))(((........-----))))))))))--------)(((.....)))....... ( -27.86, z-score = -0.61, R) >droSim1.chr3R 25464773 114 + 27517382 ---ACAAUAAUGGGUCGAAAGAACGAAAAA----------ACAAAAUACU---UUUGGGCAGUGGGAGACCGCUUACCAGGAACUAUGCUGAAAUCCCCAC-----UUCUUCGCUC--------GGCCAACGCGGCGUAUGAG ---.......((((((....))........----------.((..(((.(---(((((..(((((....)))))..))))))..)))..)).....)))).-----.((..(((((--------(.....)).))))...)). ( -31.20, z-score = -1.33, R) >consensus ___ACAAUAAUGAGUCGAAAGAACAUGAA__A________ACAAAAUACU___UU_GGGCAGUGGGAGACCGCUUACCAGGAACUAUGCUGAAAUCCCCAC_____UUUUUCACUC________GGCCAACGCGGCGUAUGAG .............................................((((.......((..(((((....)))))..))...............................................(((.....)))))))... (-11.27 = -11.94 + 0.68)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:56:01 2011