| Sequence ID | dm3.chr3R |
|---|---|
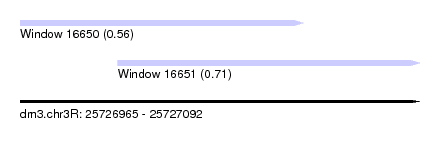
| Location | 25,726,965 – 25,727,092 |
| Length | 127 |
| Max. P | 0.711541 |

| Location | 25,726,965 – 25,727,055 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 81.05 |
| Shannon entropy | 0.32071 |
| G+C content | 0.47790 |
| Mean single sequence MFE | -22.98 |
| Consensus MFE | -17.25 |
| Energy contribution | -17.92 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.556292 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
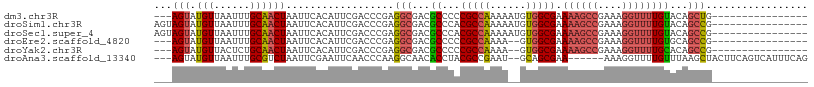
>dm3.chr3R 25726965 90 + 27905053 ---AGUAUGUUAAUUUGCAACUAAUUCACAUUCGACCCGAGGCGACGCCCCCGCCAAAAAUGUGGCGAAAAGCCGAAAGGUUUUGUACAGCUG---------------- ---(((.(((......))))))....((((((((...)))((((.......))))....)))))((..((((((....)))))).....))..---------------- ( -22.20, z-score = -1.07, R) >droSim1.chr3R 25393179 93 + 27517382 AGUAGUAUGUUAAUUUGCAACUAAUUCACAUUCGACCCGAGGCGACGCCCACGCCAAAAAUGUGGCGAAAAGCCGAAAGGUUUUGUACAGCCG---------------- ..((((.(((......))))))).................(((.((.....(((((......))))).((((((....))))))))...))).---------------- ( -25.90, z-score = -2.01, R) >droSec1.super_4 4568675 93 + 6179234 AGUAGUAUGUUAAUUUGCAACUAAUUCACAUUCGACCCGAGGCGACGCCCACGCCAAAAAUGUGGCGAAAAGCCGAAAGGUUUUGUACAGCCG---------------- ..((((.(((......))))))).................(((.((.....(((((......))))).((((((....))))))))...))).---------------- ( -25.90, z-score = -2.01, R) >droEre2.scaffold_4820 8186827 88 - 10470090 ---AGUAUGUUAAUUUGCAACUAAUUCACAUUCGACCCGAGGCGACGCCCCCGCCAAAA--GUGGCGAAAAGCCGAAAGGUUUUGUGCAGCCG---------------- ---.(.((((.((((.......)))).)))).).......(((..(((...(((((...--.))))).((((((....)))))))))..))).---------------- ( -27.00, z-score = -2.41, R) >droYak2.chr3R 8085468 88 - 28832112 ---AGUAUGUUACUCUGCAACUAAUUCACAUUCGACCCGAGGCGACGCCCCCGCCAAAA--GUGGCGAAAAGCCGAAAGGUUUUGCACAGCCG---------------- ---(((.(((......))))))..................(((...((...(((((...--.))))).((((((....))))))))...))).---------------- ( -24.50, z-score = -1.61, R) >droAna3.scaffold_13340 20718010 98 + 23697760 ---AGUAUGUUAAUUUGCGUCUAAUUCGAAUUCAACCCAAGGCAACACCUACGCCGAAU--GCAGCGAA------AAAGGUUUUGUUUAAGCUACUUCAGUCAUUUCAG ---((((.(((...(((..((......))...)))....((((((.((((.(((.....--...)))..------..)))).)))))).)))))))............. ( -12.40, z-score = 1.44, R) >consensus ___AGUAUGUUAAUUUGCAACUAAUUCACAUUCGACCCGAGGCGACGCCCACGCCAAAA__GUGGCGAAAAGCCGAAAGGUUUUGUACAGCCG________________ ...(((.(((......))))))..................(((...((...(((((......))))).((((((....))))))))...)))................. (-17.25 = -17.92 + 0.67)

| Location | 25,726,996 – 25,727,092 |
|---|---|
| Length | 96 |
| Sequences | 7 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 63.86 |
| Shannon entropy | 0.64602 |
| G+C content | 0.51872 |
| Mean single sequence MFE | -28.21 |
| Consensus MFE | -12.87 |
| Energy contribution | -13.34 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.45 |
| Mean z-score | -0.99 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.711541 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3R 25726996 96 + 27905053 ACCCGAGGCGACGCCC-----CCGCCAAAAAUGUGGCGAAAAGCCGAAAGGUUUUGUACAGCUGGCAUUAAUC-CGGCAAAAACUGGCACACACACACUUAA--------------- ......((((......-----.)))).....((((((..((((((....)))))).....(((((.......)-))))........)).)))).........--------------- ( -28.30, z-score = -1.25, R) >droSim1.chr3R 25393213 92 + 27517382 ACCCGAGGCGACGCCC-----ACGCCAAAAAUGUGGCGAAAAGCCGAAAGGUUUUGUACAGCCGGCAUUAAUC-CGGCAAAAACUGGCACACACUUAA------------------- ......((((......-----.)))).....((((((..((((((....)))))).....(((((.......)-))))........)).)))).....------------------- ( -30.10, z-score = -1.98, R) >droSec1.super_4 4568709 92 + 6179234 ACCCGAGGCGACGCCC-----ACGCCAAAAAUGUGGCGAAAAGCCGAAAGGUUUUGUACAGCCGGCAUUAAUC-CGGCAAAAACUGGCACACACUUAA------------------- ......((((......-----.)))).....((((((..((((((....)))))).....(((((.......)-))))........)).)))).....------------------- ( -30.10, z-score = -1.98, R) >droEre2.scaffold_4820 8186858 90 - 10470090 ACCCGAGGCGACGCCC-----CCGCCAAAA--GUGGCGAAAAGCCGAAAGGUUUUGUGCAGCCGGCAUUAAUC-UGGCAAAAACUGGCACACACUUAA------------------- ......((((......-----.))))..((--((((((.((((((....)))))).))).(((((.......)-)))).............)))))..------------------- ( -30.90, z-score = -1.56, R) >droYak2.chr3R 8085499 90 - 28832112 ACCCGAGGCGACGCCC-----CCGCCAAAA--GUGGCGAAAAGCCGAAAGGUUUUGCACAGCCGGCAUUAAUC-UGGCAAAAACUGGCACACACUUAA------------------- ......((((......-----.))))..((--((((((((..(((....))))))))...(((((.......)-)))).............)))))..------------------- ( -28.60, z-score = -1.24, R) >droAna3.scaffold_13340 20718041 100 + 23697760 ACCCAAGGCAACACCU-----ACGCCGAAU--GCAGCGAAA------AAGGUUUUGUUUAAGCUACUUCAGUCAUUUCAGCCAUUUAUUUACGCGUAUGUGGCACACAUUCUA---- ......(((.......-----..)))((((--(....(((.------..(((((.....)))))..)))..........(((((.(((......))).)))))...)))))..---- ( -18.70, z-score = 0.03, R) >droPer1.super_7 544088 115 - 4445127 -CCUCAGACGUCACCCGUAACCCGCCGCGAGUGCAGAGAGAAAGGCUUAGAAUUUGCGUGGGCGGUGUGCUACAAUAGAGGCAUUACUGUA-GCUCCAGAAGCUGACACACUUCGCA -..((((...(((((((...(((((.((((((.(.(((.......))).).)))))))))))))).))((((((.(((.....))).))))-))....))..))))........... ( -30.80, z-score = 1.02, R) >consensus ACCCGAGGCGACGCCC_____CCGCCAAAA_UGUGGCGAAAAGCCGAAAGGUUUUGUACAGCCGGCAUUAAUC_CGGCAAAAACUGGCACACACUUAA___________________ ......((((............))))......((((((((..(((....))))))))...(((((..................)))))...)))....................... (-12.87 = -13.34 + 0.48)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:55:56 2011