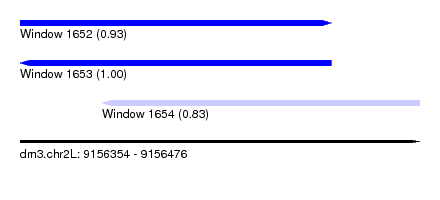
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 9,156,354 – 9,156,476 |
| Length | 122 |
| Max. P | 0.999454 |

| Location | 9,156,354 – 9,156,449 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 63.80 |
| Shannon entropy | 0.67320 |
| G+C content | 0.40334 |
| Mean single sequence MFE | -21.79 |
| Consensus MFE | -8.74 |
| Energy contribution | -9.18 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.933661 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
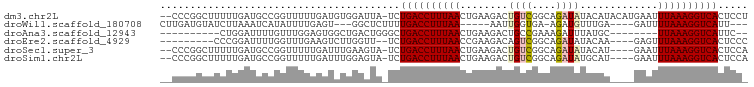
>dm3.chr2L 9156354 95 + 23011544 --CCCGGCUUUUUGAUGCCGGUUUUUGAUGUGGAUUA-UCUGACCUUUAACUGAAGACUGUCGGCAGAUAUACAUACAUGAAUUUAAAGGUCACUCCU --.(((((........)))))..........(((...-..(((((((((((((..(.....)..))).....((....))...)))))))))).))). ( -24.10, z-score = -1.90, R) >droWil1.scaffold_180708 106366 82 + 12563649 CUUGAUGUAUCUUAAAUCAUAUUUUGAGU---GGCUCUUUUGACCUUUAA-----AAUUGGUGA-AGAUGUUUGA----GAUUUUAAAGGUCAUU--- ...((.((..((((((......)))))).---.))))...((((((((((-----((((..(((-(....)))).----))))))))))))))..--- ( -19.60, z-score = -2.57, R) >droAna3.scaffold_12943 503960 78 + 5039921 ----------CUGGAUUUUGUUUGGAGUGGCUGACUGGGCUGACCUUUAACUGAAGACUGCCGAAAGAUUUAUGC--------UUAAAGGUCAUUC-- ----------(..(((...)))..)...((((.....))))((((((((((((((..((......)).)))).).--------)))))))))....-- ( -15.70, z-score = -0.07, R) >droEre2.scaffold_4929 9761563 83 + 26641161 ---------CCCGGAUUUUGGUUUGAAGUCUUGGUU--UCUGACCUUUAACCGAAGACAGUCGGCAGAUAUACAA----GAGUUUAAAGGUCACUCCC ---------((.(((((((.....))))))).))..--..((((((((((((((......)))).......((..----..))))))))))))..... ( -20.20, z-score = -0.47, R) >droSec1.super_3 4622084 91 + 7220098 --CCCGGCUUUUUGAUGCCGGUUUUUGAUUUGAAGUA-UCUGACCUUUAACUGAAGACUGUCGGCAGAUAUACAU----GAAUUUAAAGGUCACUCCA --.(((((........)))))................-..((((((((((........((((....)))).....----....))))))))))..... ( -22.33, z-score = -1.72, R) >droSim1.chr2L 8936407 91 + 22036055 --CCCGGCUUUUUGAUGCCGGUUUUUGAUUUGGAGUA-UCUGACCUUUAACUGAAGACUGUCGGCAGAUAUGCAU----GAAUUUAAAGGUCACUCCA --.(((((........))))).........((((((.-...(((((((((.(((......)))(((....)))..----....))))))))))))))) ( -28.80, z-score = -3.13, R) >consensus __CCCGGCUUCUUGAUGCCGGUUUUUGAUGUGGACUA_UCUGACCUUUAACUGAAGACUGUCGGCAGAUAUACAU____GAAUUUAAAGGUCACUCC_ ........................................((((((((((........((((....)))).............))))))))))..... ( -8.74 = -9.18 + 0.44)
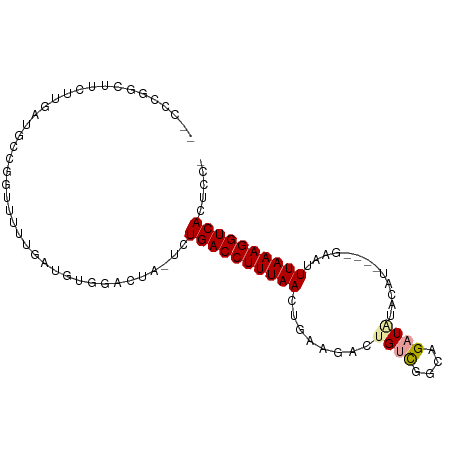
| Location | 9,156,354 – 9,156,449 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 63.80 |
| Shannon entropy | 0.67320 |
| G+C content | 0.40334 |
| Mean single sequence MFE | -20.86 |
| Consensus MFE | -9.34 |
| Energy contribution | -9.15 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.90 |
| SVM RNA-class probability | 0.999454 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 9156354 95 - 23011544 AGGAGUGACCUUUAAAUUCAUGUAUGUAUAUCUGCCGACAGUCUUCAGUUAAAGGUCAGA-UAAUCCACAUCAAAAACCGGCAUCAAAAAGCCGGG-- .(((.((((((((((.....(((..(((....)))..)))........))))))))))..-...)))..........(((((........))))).-- ( -26.52, z-score = -3.59, R) >droWil1.scaffold_180708 106366 82 - 12563649 ---AAUGACCUUUAAAAUC----UCAAACAUCU-UCACCAAUU-----UUAAAGGUCAAAAGAGCC---ACUCAAAAUAUGAUUUAAGAUACAUCAAG ---..(((((((((((((.----..........-......)))-----))))))))))...(((..---.)))......................... ( -13.43, z-score = -3.44, R) >droAna3.scaffold_12943 503960 78 - 5039921 --GAAUGACCUUUAA--------GCAUAAAUCUUUCGGCAGUCUUCAGUUAAAGGUCAGCCCAGUCAGCCACUCCAAACAAAAUCCAG---------- --...((((((((((--------((............)).(....)..))))))))))..............................---------- ( -10.40, z-score = -0.80, R) >droEre2.scaffold_4929 9761563 83 - 26641161 GGGAGUGACCUUUAAACUC----UUGUAUAUCUGCCGACUGUCUUCGGUUAAAGGUCAGA--AACCAAGACUUCAAACCAAAAUCCGGG--------- .(((.(((((((((.((..----..))......(((((......))))))))))))))((--(........))).........)))...--------- ( -19.40, z-score = -0.71, R) >droSec1.super_3 4622084 91 - 7220098 UGGAGUGACCUUUAAAUUC----AUGUAUAUCUGCCGACAGUCUUCAGUUAAAGGUCAGA-UACUUCAAAUCAAAAACCGGCAUCAAAAAGCCGGG-- (((((((((((((((....----.((.....(((....)))....)).)))))))))...-.)))))).........(((((........))))).-- ( -26.60, z-score = -3.99, R) >droSim1.chr2L 8936407 91 - 22036055 UGGAGUGACCUUUAAAUUC----AUGCAUAUCUGCCGACAGUCUUCAGUUAAAGGUCAGA-UACUCCAAAUCAAAAACCGGCAUCAAAAAGCCGGG-- (((((((((((((((....----.((.....(((....)))....)).)))))))))...-.)))))).........(((((........))))).-- ( -28.80, z-score = -4.53, R) >consensus _GGAGUGACCUUUAAAUUC____AUGUAUAUCUGCCGACAGUCUUCAGUUAAAGGUCAGA_UAAUCCAAAUCAAAAACCGGAAUCAAAAAGCCGGG__ .....((((((((((..........(((....))).............))))))))))........................................ ( -9.34 = -9.15 + -0.19)
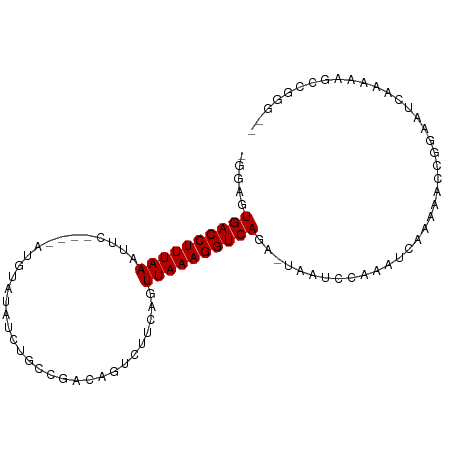
| Location | 9,156,379 – 9,156,476 |
|---|---|
| Length | 97 |
| Sequences | 7 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 70.79 |
| Shannon entropy | 0.58606 |
| G+C content | 0.39071 |
| Mean single sequence MFE | -16.35 |
| Consensus MFE | -8.65 |
| Energy contribution | -8.80 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.827345 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 9156379 97 - 23011544 AGUCAGUACAAAUCCGUUCACAAUUGUAGGAGUGACCUUUAAAUUCAUGUAUGUAUAUCUGCCGACAGUCUUCAGUUAAAGGUCAGAUAAUCCACAU .(((........(((...((....))..))).((((((((((.....(((..(((....)))..)))........)))))))))))))......... ( -17.72, z-score = -0.95, R) >droWil1.scaffold_180708 106391 79 - 12563649 -UUCAAUAAAAAACUUUAUGCAA-------AAUGACCUUUAAAAUCUCA----AACAUCU-UCACCAAUU-----UUAAAGGUCAAAAGAGCCACUC -......................-------..(((((((((((((....----.......-......)))-----))))))))))............ ( -12.73, z-score = -3.96, R) >droAna3.scaffold_12943 503983 81 - 5039921 UGUCAGUUCAAAUCCGUUCACAACUC---GAAUGACCUUUAAGCA--------UAAAUCUUUCGGCAGUCUUCAGUUAAAGGUCAGCCCAGU----- ......................(((.---(..((((((((((((.--------...........)).(....)..))))))))))..).)))----- ( -12.20, z-score = -0.91, R) >droEre2.scaffold_4929 9761586 87 - 26641161 AGUCAGUACAAAUCCGUUCACAACUGGGGGAGUGACCUUUAAACUCUUG----UAUAUCUGCCGACUGUCUUCGGUUAAAGGUCAGAAACC------ ..((........(((.((((....))))))).(((((((((.((....)----)......(((((......))))))))))))))))....------ ( -19.40, z-score = 0.09, R) >droYak2.chr2L 11823871 81 - 22324452 AGUCAGUACAAAUCCGUUCACAACUCGAGGAGUGACCUUUACAUUCAUG----UCUAUCUGCCGAAAGUCUUCAGUUAAAGGUCA------------ ............(((.............))).(((((((((.(((...(----.((.((....)).)).)...))))))))))))------------ ( -13.02, z-score = 0.28, R) >droSec1.super_3 4622109 93 - 7220098 AGUCAGUACGAAUCCGUUCACAAUUUGUGGAGUGACCUUUAAAUUCAUG----UAUAUCUGCCGACAGUCUUCAGUUAAAGGUCAGAUACUUCAAAU .(((............(((((.....))))).((((((((((.....((----.....(((....)))....)).)))))))))))))......... ( -19.10, z-score = -1.20, R) >droSim1.chr2L 8936432 93 - 22036055 AGUCAGUACGAAUCCGUUCACAAUUGGUGGAGUGACCUUUAAAUUCAUG----CAUAUCUGCCGACAGUCUUCAGUUAAAGGUCAGAUACUCCAAAU ..((((((((....))).....)))))(((((((((((((((.....((----.....(((....)))....)).)))))))))....))))))... ( -20.30, z-score = -1.12, R) >consensus AGUCAGUACAAAUCCGUUCACAACUGG_GGAGUGACCUUUAAAUUCAUG____UAUAUCUGCCGACAGUCUUCAGUUAAAGGUCAGAUAAUCCA___ ................................((((((((((.................................))))))))))............ ( -8.65 = -8.80 + 0.14)

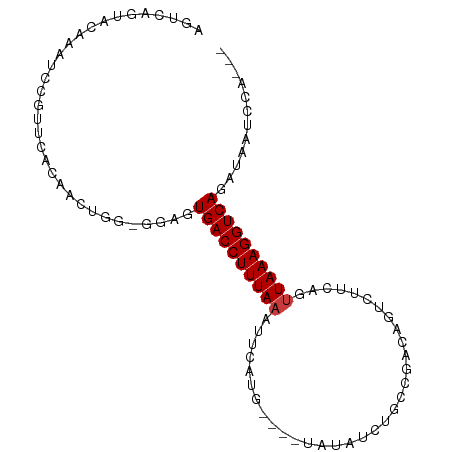
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:27:40 2011