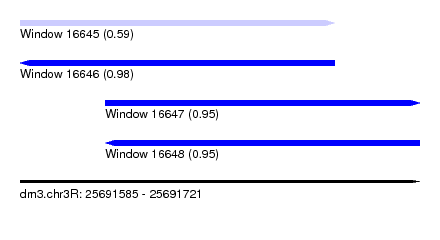
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 25,691,585 – 25,691,721 |
| Length | 136 |
| Max. P | 0.978169 |

| Location | 25,691,585 – 25,691,692 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 91.48 |
| Shannon entropy | 0.14992 |
| G+C content | 0.29940 |
| Mean single sequence MFE | -23.36 |
| Consensus MFE | -19.94 |
| Energy contribution | -20.26 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.585694 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3R 25691585 107 + 27905053 AAUGCAGGUACUUAAAUGAUAAAUAUAUAUUUGCCAAUAUGGCUUGUUCGAGUUCGAGCAAUAAUUAAUUGGCUUUUUCCAUAAUUAAUAAGCAAUUUAGAUGCAGU ..((((((((..((.(((.....))).))..))))......(((((((((....)))))..((((((..(((......)))))))))..))))........)))).. ( -20.20, z-score = -0.19, R) >droEre2.scaffold_4820 8160794 105 - 10470090 AAUGCAGGUACUUAAAUGAU--UAAUAUCUUUGCCAAUUUCGAUUGUUCGAGUUCGAGCAAUAAUUAAUUGGCUUUUUCUAUAAUUUAUAAGCAAUUUAGAUGUAGU ..((((((((.((((....)--))))))))..(((((((...((((((((....))))))))....)))))))..................)))............. ( -23.80, z-score = -2.36, R) >droYak2.chr3R 8058817 105 - 28832112 AAUGCAGGUACUUAAAUGAU--UUAUAUCUUUGCCAAUUUUGUUUGCUCGCGUUCGAGCAAUAAUUAAUUGGCUUUUUCUAUAAUUCAUAAGCAAUUUAGACGCAGU ..(((..........((((.--(((((.....(((((((....(((((((....))))))).....)))))))......))))).))))((....)).....))).. ( -24.60, z-score = -2.14, R) >droSec1.super_4 4542599 107 + 6179234 AAUGCAGGUACUUAAAUGAUAAAUAUAUCUUUGCCAAUUUGGCUUGCUCGAGUUCGAGCAGUAAUUAAUUGGCUUUUUCCAUAAUUUAUAAGCAAUUAAGAUGAAGU ..(((..(((.......((((....))))...(((((((..(((.(((((....))))))))....))))))).............)))..)))............. ( -24.10, z-score = -1.49, R) >droSim1.chr3R 25367066 107 + 27517382 AAUGCAGGUACUUAAAUGAUAAAUAUAUCUUUGCCAAUUUGGCUUGCUCGAGUUCGAGCAGUAAUUAAUUGGCUUUUUCCAUAAUUUAUAAGCAAUUAAGAUGAAGU ..(((..(((.......((((....))))...(((((((..(((.(((((....))))))))....))))))).............)))..)))............. ( -24.10, z-score = -1.49, R) >consensus AAUGCAGGUACUUAAAUGAUAAAUAUAUCUUUGCCAAUUUGGCUUGCUCGAGUUCGAGCAAUAAUUAAUUGGCUUUUUCCAUAAUUUAUAAGCAAUUUAGAUGAAGU ..(((............((((....))))...(((((((....(((((((....))))))).....)))))))..................)))............. (-19.94 = -20.26 + 0.32)

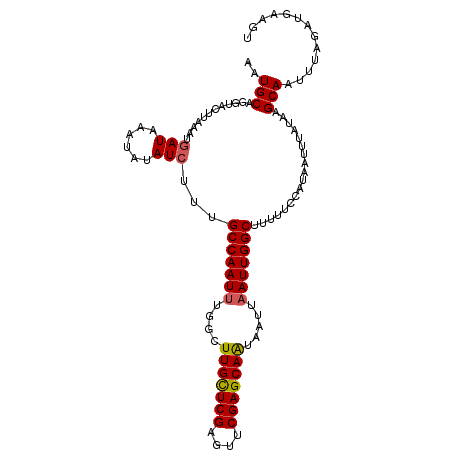
| Location | 25,691,585 – 25,691,692 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 91.48 |
| Shannon entropy | 0.14992 |
| G+C content | 0.29940 |
| Mean single sequence MFE | -22.14 |
| Consensus MFE | -19.12 |
| Energy contribution | -20.72 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.978169 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3R 25691585 107 - 27905053 ACUGCAUCUAAAUUGCUUAUUAAUUAUGGAAAAAGCCAAUUAAUUAUUGCUCGAACUCGAACAAGCCAUAUUGGCAAAUAUAUAUUUAUCAUUUAAGUACCUGCAUU ..(((((((((((((.....))))).))))....((((((......(((.(((....))).))).....)))))).......((((((.....))))))..)))).. ( -19.20, z-score = -1.28, R) >droEre2.scaffold_4820 8160794 105 + 10470090 ACUACAUCUAAAUUGCUUAUAAAUUAUAGAAAAAGCCAAUUAAUUAUUGCUCGAACUCGAACAAUCGAAAUUGGCAAAGAUAUUA--AUCAUUUAAGUACCUGCAUU .............((((((...............(((((((....((((.(((....))).))))...)))))))...(((....--)))...))))))........ ( -17.80, z-score = -2.25, R) >droYak2.chr3R 8058817 105 + 28832112 ACUGCGUCUAAAUUGCUUAUGAAUUAUAGAAAAAGCCAAUUAAUUAUUGCUCGAACGCGAGCAAACAAAAUUGGCAAAGAUAUAA--AUCAUUUAAGUACCUGCAUU ..((((.......(((((((((.(((((......(((((((.....(((((((....)))))))....))))))).....)))))--.)))..))))))..)))).. ( -25.90, z-score = -3.42, R) >droSec1.super_4 4542599 107 - 6179234 ACUUCAUCUUAAUUGCUUAUAAAUUAUGGAAAAAGCCAAUUAAUUACUGCUCGAACUCGAGCAAGCCAAAUUGGCAAAGAUAUAUUUAUCAUUUAAGUACCUGCAUU .............((((((...............(((((((......((((((....)))))).....)))))))...((((....))))...))))))........ ( -23.90, z-score = -3.02, R) >droSim1.chr3R 25367066 107 - 27517382 ACUUCAUCUUAAUUGCUUAUAAAUUAUGGAAAAAGCCAAUUAAUUACUGCUCGAACUCGAGCAAGCCAAAUUGGCAAAGAUAUAUUUAUCAUUUAAGUACCUGCAUU .............((((((...............(((((((......((((((....)))))).....)))))))...((((....))))...))))))........ ( -23.90, z-score = -3.02, R) >consensus ACUGCAUCUAAAUUGCUUAUAAAUUAUGGAAAAAGCCAAUUAAUUAUUGCUCGAACUCGAGCAAGCCAAAUUGGCAAAGAUAUAUUUAUCAUUUAAGUACCUGCAUU .............((((((...............(((((((.....(((((((....)))))))....)))))))...((((....))))...))))))........ (-19.12 = -20.72 + 1.60)

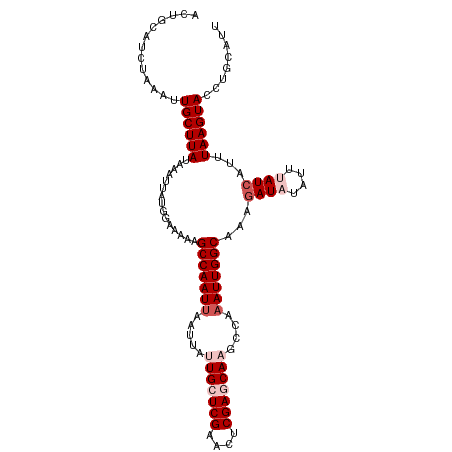
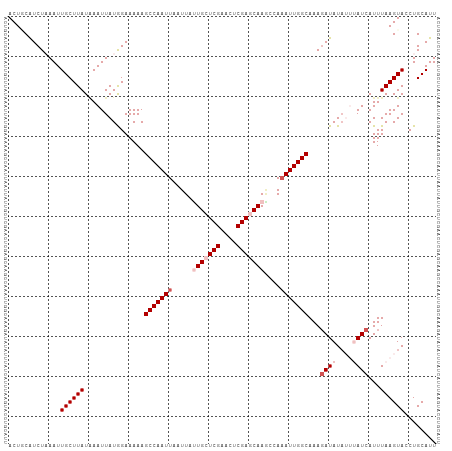
| Location | 25,691,614 – 25,691,721 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 89.18 |
| Shannon entropy | 0.19233 |
| G+C content | 0.33273 |
| Mean single sequence MFE | -27.96 |
| Consensus MFE | -23.48 |
| Energy contribution | -23.84 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.951343 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 25691614 107 + 27905053 UUUGCCAAUAUGGCUUGUUCGAGUUCGAGCAAUAAUUAAUUGGCUUUUUCCAUAAUUAAUAAGCAAUUUAGAUGCAGUUAG----UUAGUUUGAAUGUGAUUGCUUUCCAG ...((((((.....(((((((....)))))))......))))))................((((((((.....(((.((((----.....)))).)))))))))))..... ( -22.80, z-score = -1.04, R) >droEre2.scaffold_4820 8160821 111 - 10470090 UUUGCCAAUUUCGAUUGUUCGAGUUCGAGCAAUAAUUAAUUGGCUUUUUCUAUAAUUUAUAAGCAAUUUAGAUGUAGUUAUAUGCUUAGUUUGAAUGUGAUUGCUAUUCUG ...(((((((...((((((((....))))))))....)))))))........................((((.((((((((((.(.......).))))))))))...)))) ( -28.60, z-score = -3.21, R) >droYak2.chr3R 8058844 111 - 28832112 UUUGCCAAUUUUGUUUGCUCGCGUUCGAGCAAUAAUUAAUUGGCUUUUUCUAUAAUUCAUAAGCAAUUUAGACGCAGUUAUAUGCUUAGUUUGAAUGUGAUUGCUACUCGG ...(((((((....(((((((....))))))).....)))))))....((((.(((((....).)))))))).((((((((((.(.......).))))))))))....... ( -28.50, z-score = -2.32, R) >droSec1.super_4 4542628 111 + 6179234 UUUGCCAAUUUGGCUUGCUCGAGUUCGAGCAGUAAUUAAUUGGCUUUUUCCAUAAUUUAUAAGCAAUUAAGAUGAAGUCAUGUGCUUAGUUUGAAUGUGAUUGCUUUCCAG ...(((((((..(((.(((((....))))))))....)))))))................(((((((((((((.((((.....)))).)))).....)))))))))..... ( -29.10, z-score = -2.12, R) >droSim1.chr3R 25367095 110 + 27517382 UUUGCCAAUUUGGCUUGCUCGAGUUCGAGCAGUAAUUAAUUGGCUUUUUCCAUAAUUUAUAAGCAAUUAAGAUGAAGUCAUG-GCUUAGUUUGAAUGUGAUUGCUUUCCAG ...(((((((..(((.(((((....))))))))....)))))))................(((((((((((((.((((....-)))).)))).....)))))))))..... ( -30.80, z-score = -2.56, R) >consensus UUUGCCAAUUUGGCUUGCUCGAGUUCGAGCAAUAAUUAAUUGGCUUUUUCCAUAAUUUAUAAGCAAUUUAGAUGAAGUUAUAUGCUUAGUUUGAAUGUGAUUGCUUUCCAG ...(((((((....(((((((....))))))).....))))))).................((((((((((((..(((.....)))..))))).....)))))))...... (-23.48 = -23.84 + 0.36)

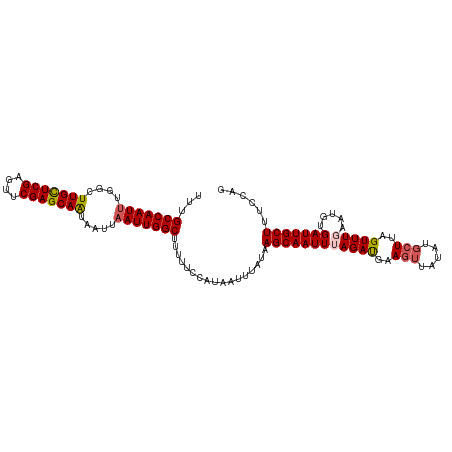
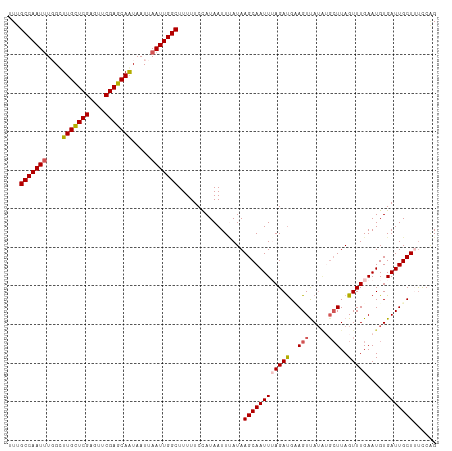
| Location | 25,691,614 – 25,691,721 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 89.18 |
| Shannon entropy | 0.19233 |
| G+C content | 0.33273 |
| Mean single sequence MFE | -21.33 |
| Consensus MFE | -18.03 |
| Energy contribution | -19.43 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.949292 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 25691614 107 - 27905053 CUGGAAAGCAAUCACAUUCAAACUAA----CUAACUGCAUCUAAAUUGCUUAUUAAUUAUGGAAAAAGCCAAUUAAUUAUUGCUCGAACUCGAACAAGCCAUAUUGGCAAA .....(((((((..............----..............)))))))................((((((......(((.(((....))).))).....))))))... ( -16.79, z-score = -1.09, R) >droEre2.scaffold_4820 8160821 111 + 10470090 CAGAAUAGCAAUCACAUUCAAACUAAGCAUAUAACUACAUCUAAAUUGCUUAUAAAUUAUAGAAAAAGCCAAUUAAUUAUUGCUCGAACUCGAACAAUCGAAAUUGGCAAA ..((((.(......)))))....((((((.................))))))...............(((((((....((((.(((....))).))))...)))))))... ( -18.13, z-score = -2.55, R) >droYak2.chr3R 8058844 111 + 28832112 CCGAGUAGCAAUCACAUUCAAACUAAGCAUAUAACUGCGUCUAAAUUGCUUAUGAAUUAUAGAAAAAGCCAAUUAAUUAUUGCUCGAACGCGAGCAAACAAAAUUGGCAAA ...............(((((...((((((.................)))))))))))..........(((((((.....(((((((....)))))))....)))))))... ( -24.33, z-score = -2.49, R) >droSec1.super_4 4542628 111 - 6179234 CUGGAAAGCAAUCACAUUCAAACUAAGCACAUGACUUCAUCUUAAUUGCUUAUAAAUUAUGGAAAAAGCCAAUUAAUUACUGCUCGAACUCGAGCAAGCCAAAUUGGCAAA ...........((.(((......((((((.(((....)))......))))))......)))))....(((((((......((((((....)))))).....)))))))... ( -23.70, z-score = -2.49, R) >droSim1.chr3R 25367095 110 - 27517382 CUGGAAAGCAAUCACAUUCAAACUAAGC-CAUGACUUCAUCUUAAUUGCUUAUAAAUUAUGGAAAAAGCCAAUUAAUUACUGCUCGAACUCGAGCAAGCCAAAUUGGCAAA .....(((((((...........((((.-.(((....))))))))))))))................(((((((......((((((....)))))).....)))))))... ( -23.70, z-score = -2.32, R) >consensus CUGGAAAGCAAUCACAUUCAAACUAAGCACAUAACUGCAUCUAAAUUGCUUAUAAAUUAUGGAAAAAGCCAAUUAAUUAUUGCUCGAACUCGAGCAAGCCAAAUUGGCAAA .....(((((((................................)))))))................(((((((.....(((((((....)))))))....)))))))... (-18.03 = -19.43 + 1.40)

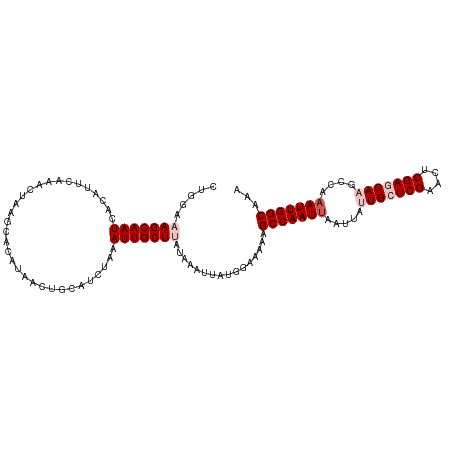
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:55:54 2011