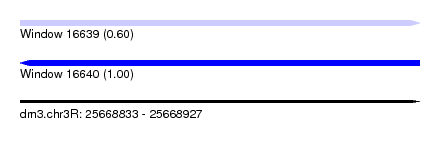
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 25,668,833 – 25,668,927 |
| Length | 94 |
| Max. P | 0.999078 |

| Location | 25,668,833 – 25,668,927 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 66.11 |
| Shannon entropy | 0.50184 |
| G+C content | 0.28026 |
| Mean single sequence MFE | -14.85 |
| Consensus MFE | -7.82 |
| Energy contribution | -8.45 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.600173 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 25668833 94 + 27905053 UUUGGCCAUCAUCUAAAUUCAAAGAUUUGUUUUUAAUUAUU---CUAAACUCAACUUAUUUUGGCCAAUUAAGACAAUCAUAAUGGUAGAAUGUUUA--------------- .(((((((..((((........))))(((..((((......---.))))..))).......)))))))...(((((.((.........)).))))).--------------- ( -14.10, z-score = -0.88, R) >droSim1.chr3R 25343150 78 + 27517382 UUUGGCCAGCAUCUAAAUUCAGAGAUUUGUUUUUAAUUAUU---CUAAAUUCAACUUACUUUGGCCAAUUAAGAUUGUU-UA------------------------------ .((((((((.((((........))))(((..((((......---.))))..)))......))))))))...........-..------------------------------ ( -13.00, z-score = -1.26, R) >droSec1.super_4 4519218 78 + 6179234 UUUGGCCAGCAUCUAAAUUCAGAGAUUUGUUUUUAAUUAUU---CUAAAUUCAACUUACUUUGGCCAAUUAAGAUUGUU-UA------------------------------ .((((((((.((((........))))(((..((((......---.))))..)))......))))))))...........-..------------------------------ ( -13.00, z-score = -1.26, R) >droEre2.scaffold_4820 8136474 110 - 10470090 UUUGGCCAGCAUCUUAAUUCAGCGACUUAUGUUUAAUAUUUGCACUUAACUUA-UUUACUUUGGCCA-UUAAAACGAUGGCCAUUUAAGACCAUCAUACGAGAGAUUAUUUA ..(((......((((((....((((..(((.....))).))))..........-.......((((((-((.....)))))))).))))))))).....(....)........ ( -19.30, z-score = -1.54, R) >consensus UUUGGCCAGCAUCUAAAUUCAGAGAUUUGUUUUUAAUUAUU___CUAAACUCAACUUACUUUGGCCAAUUAAGACUAUU_UA______________________________ .(((((((...(((........))).(((..((((..........))))..))).......)))))))............................................ ( -7.82 = -8.45 + 0.63)

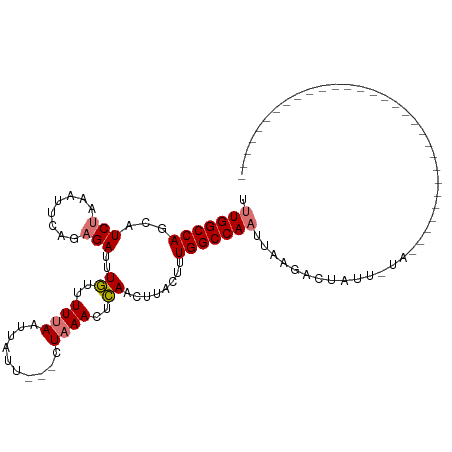
| Location | 25,668,833 – 25,668,927 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 66.11 |
| Shannon entropy | 0.50184 |
| G+C content | 0.28026 |
| Mean single sequence MFE | -19.66 |
| Consensus MFE | -12.62 |
| Energy contribution | -12.75 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.63 |
| SVM RNA-class probability | 0.999078 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 25668833 94 - 27905053 ---------------UAAACAUUCUACCAUUAUGAUUGUCUUAAUUGGCCAAAAUAAGUUGAGUUUAG---AAUAAUUAAAAACAAAUCUUUGAAUUUAGAUGAUGGCCAAA ---------------...(((.((.........)).))).....(((((((.......((((((((((---(.................)))))))))))....))))))). ( -15.73, z-score = -1.29, R) >droSim1.chr3R 25343150 78 - 27517382 ------------------------------UA-AACAAUCUUAAUUGGCCAAAGUAAGUUGAAUUUAG---AAUAAUUAAAAACAAAUCUCUGAAUUUAGAUGCUGGCCAAA ------------------------------..-...........(((((((..(((..((((((((((---(.................))))))))))).)))))))))). ( -18.83, z-score = -3.87, R) >droSec1.super_4 4519218 78 - 6179234 ------------------------------UA-AACAAUCUUAAUUGGCCAAAGUAAGUUGAAUUUAG---AAUAAUUAAAAACAAAUCUCUGAAUUUAGAUGCUGGCCAAA ------------------------------..-...........(((((((..(((..((((((((((---(.................))))))))))).)))))))))). ( -18.83, z-score = -3.87, R) >droEre2.scaffold_4820 8136474 110 + 10470090 UAAAUAAUCUCUCGUAUGAUGGUCUUAAAUGGCCAUCGUUUUAA-UGGCCAAAGUAAA-UAAGUUAAGUGCAAAUAUUAAACAUAAGUCGCUGAAUUAAGAUGCUGGCCAAA ...............(((((((((......))))))))).....-((((((..(((..-(.((((.(((((...............).)))).)))).)..))))))))).. ( -25.26, z-score = -2.48, R) >consensus ______________________________UA_AACAAUCUUAAUUGGCCAAAGUAAGUUGAAUUUAG___AAUAAUUAAAAACAAAUCUCUGAAUUUAGAUGCUGGCCAAA ............................................(((((((..((....(((((((((......................))))))))).))..))))))). (-12.62 = -12.75 + 0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:55:47 2011