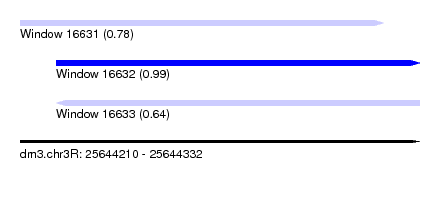
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 25,644,210 – 25,644,332 |
| Length | 122 |
| Max. P | 0.990554 |

| Location | 25,644,210 – 25,644,321 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 89.26 |
| Shannon entropy | 0.16700 |
| G+C content | 0.51838 |
| Mean single sequence MFE | -33.45 |
| Consensus MFE | -29.50 |
| Energy contribution | -29.62 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.778762 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
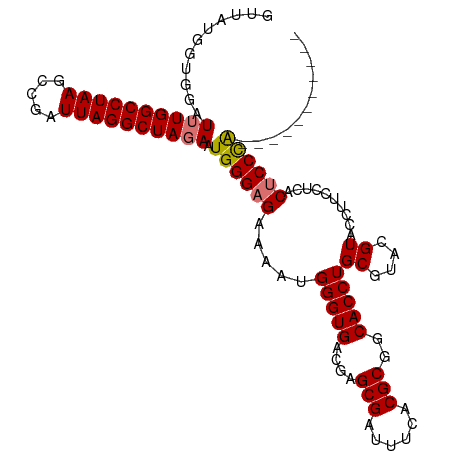
>dm3.chr3R 25644210 111 + 27905053 GUUAUGGUGGAUUUGGCCUAAGCCGAUUAGGCUAGAAUGGGAGAAAAUGGGUGACGAGCGAUUUCACGCGACACCUGCGUACGUACCUUUCUCACUCCCAACUCCCAACUC ....(((..(.((((((((((.....)))))))))).((((((.....(((((.((...(....)(((((.....))))).)))))))......)))))).)..))).... ( -37.40, z-score = -2.54, R) >droYak2.chr3R 8007734 100 - 28832112 GUUAUGGUGGAUUUGGCCUAAGCCGAUUAGGCUAGAAUGGGAGAAAAUGGGUGACGAGCGAUUUCACGCGGCACCUGCGUACGUAUUGUACAUACACCUG----------- .....((((..((((((((((.....))))))))))............(((((....(((......)))..)))))..((((.....))))...))))..----------- ( -31.70, z-score = -1.81, R) >droSec1.super_4 4494747 100 + 6179234 GUUAUGGUGGAUUUGGCCUAAGCCGAUUAGGCUAGAAUGGGAGAAAAUGGGUGACGAGCGAUUUCACGCGGCACCUGCGUACGUACCUUCCUCACUCCCA----------- ...........((((((((((.....)))))))))).((((((.....(((((.((...(....)((((((...)))))).)))))))......))))))----------- ( -33.50, z-score = -1.30, R) >droSim1.chr3R 25316795 100 + 27517382 GUUAUGGUGGAUUUGGCCUAAGCCGAUUAGGCUACAAUGGGAGAAAAUGGGUGACGAGCGAUUUCACGCGGCACCUGCGUACGUACCUUCCUCACUCCCA----------- .............((((((((.....))))))))...((((((.....(((((.((...(....)((((((...)))))).)))))))......))))))----------- ( -31.20, z-score = -0.70, R) >consensus GUUAUGGUGGAUUUGGCCUAAGCCGAUUAGGCUAGAAUGGGAGAAAAUGGGUGACGAGCGAUUUCACGCGGCACCUGCGUACGUACCUUCCUCACUCCCA___________ ...........((((((((((.....)))))))))).((((((.....(((((....(((......)))..)))))((....))..........))))))........... (-29.50 = -29.62 + 0.12)

| Location | 25,644,221 – 25,644,332 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 83.17 |
| Shannon entropy | 0.25980 |
| G+C content | 0.52508 |
| Mean single sequence MFE | -32.09 |
| Consensus MFE | -29.60 |
| Energy contribution | -29.72 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.43 |
| SVM RNA-class probability | 0.990554 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
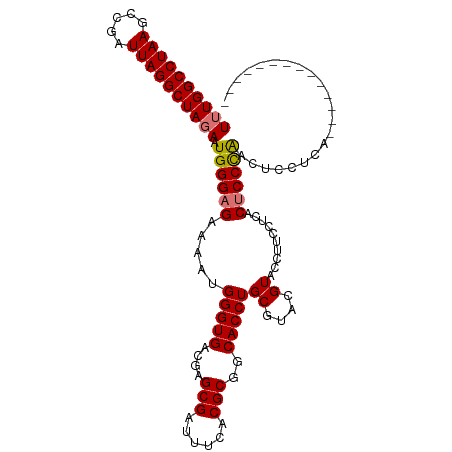
>dm3.chr3R 25644221 111 + 27905053 UUUGGCCUAAGCCGAUUAGGCUAGAAUGGGAGAAAAUGGGUGACGAGCGAUUUCACGCGACACCUGCGUACGUACCUUUCUCACUCCCAACUCCCAACUCCACACUCCUCA ((((((((((.....)))))))))).((((((.....((((((.(((.......(((((.....)))))........)))))))))....))))))............... ( -33.76, z-score = -2.63, R) >droYak2.chr3R 8007745 89 - 28832112 UUUGGCCUAAGCCGAUUAGGCUAGAAUGGGAGAAAAUGGGUGACGAGCGAUUUCACGCGGCACCUGCGUACGUAUUGUACAUACACCUG---------------------- ((((((((((.....))))))))))..(((.......(((((....(((......)))..)))))..((((.....)))).....))).---------------------- ( -28.60, z-score = -2.13, R) >droSec1.super_4 4494758 97 + 6179234 UUUGGCCUAAGCCGAUUAGGCUAGAAUGGGAGAAAAUGGGUGACGAGCGAUUUCACGCGGCACCUGCGUACGUACCUUCCUCACUCCCAACUCCUCA-------------- ((((((((((.....)))))))))).((((((.....(((((.((...(....)((((((...)))))).)))))))......))))))........-------------- ( -34.00, z-score = -2.43, R) >droSim1.chr3R 25316806 97 + 27517382 UUUGGCCUAAGCCGAUUAGGCUACAAUGGGAGAAAAUGGGUGACGAGCGAUUUCACGCGGCACCUGCGUACGUACCUUCCUCACUCCCAACUCCUCA-------------- ..((((((((.....))))))))...((((((.....(((((.((...(....)((((((...)))))).)))))))......))))))........-------------- ( -32.00, z-score = -2.13, R) >consensus UUUGGCCUAAGCCGAUUAGGCUAGAAUGGGAGAAAAUGGGUGACGAGCGAUUUCACGCGGCACCUGCGUACGUACCUUCCUCACUCCCAACUCCUCA______________ ((((((((((.....)))))))))).((((((.....(((((....(((......)))..)))))((....))..........))))))...................... (-29.60 = -29.72 + 0.12)

| Location | 25,644,221 – 25,644,332 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 83.17 |
| Shannon entropy | 0.25980 |
| G+C content | 0.52508 |
| Mean single sequence MFE | -33.92 |
| Consensus MFE | -22.86 |
| Energy contribution | -23.67 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.640889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 25644221 111 - 27905053 UGAGGAGUGUGGAGUUGGGAGUUGGGAGUGAGAAAGGUACGUACGCAGGUGUCGCGUGAAAUCGCUCGUCACCCAUUUUCUCCCAUUCUAGCCUAAUCGGCUUAGGCCAAA .((..((..((((((.(((((.((((.(((((...(((...(((((.......)))))..))).)))).).))))....)))))))))))..))..))(((....)))... ( -41.40, z-score = -2.37, R) >droYak2.chr3R 8007745 89 + 28832112 ----------------------CAGGUGUAUGUACAAUACGUACGCAGGUGCCGCGUGAAAUCGCUCGUCACCCAUUUUCUCCCAUUCUAGCCUAAUCGGCUUAGGCCAAA ----------------------...(((((((((...))))))))).((((.((.(((....))).)).)))).................((((((.....)))))).... ( -26.90, z-score = -2.61, R) >droSec1.super_4 4494758 97 - 6179234 --------------UGAGGAGUUGGGAGUGAGGAAGGUACGUACGCAGGUGCCGCGUGAAAUCGCUCGUCACCCAUUUUCUCCCAUUCUAGCCUAAUCGGCUUAGGCCAAA --------------((.((((...((.((((.((.(((((........)))))(((......))))).)))))).....)))))).....((((((.....)))))).... ( -33.70, z-score = -1.42, R) >droSim1.chr3R 25316806 97 - 27517382 --------------UGAGGAGUUGGGAGUGAGGAAGGUACGUACGCAGGUGCCGCGUGAAAUCGCUCGUCACCCAUUUUCUCCCAUUGUAGCCUAAUCGGCUUAGGCCAAA --------------((.((((...((.((((.((.(((((........)))))(((......))))).)))))).....)))))).....((((((.....)))))).... ( -33.70, z-score = -1.41, R) >consensus ______________UGAGGAGUUGGGAGUGAGGAAGGUACGUACGCAGGUGCCGCGUGAAAUCGCUCGUCACCCAUUUUCUCCCAUUCUAGCCUAAUCGGCUUAGGCCAAA ......................((((((.(((........(....).((((.((.(((....))).)).))))..))).)))))).....((((((.....)))))).... (-22.86 = -23.67 + 0.81)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:55:41 2011