| Sequence ID | dm3.chr3R |
|---|---|
| Location | 25,614,379 – 25,614,431 |
| Length | 52 |
| Max. P | 0.538211 |

| Location | 25,614,379 – 25,614,431 |
|---|---|
| Length | 52 |
| Sequences | 10 |
| Columns | 56 |
| Reading direction | reverse |
| Mean pairwise identity | 87.63 |
| Shannon entropy | 0.23706 |
| G+C content | 0.43055 |
| Mean single sequence MFE | -13.57 |
| Consensus MFE | -5.27 |
| Energy contribution | -5.06 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.25 |
| Mean z-score | -3.05 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.538211 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
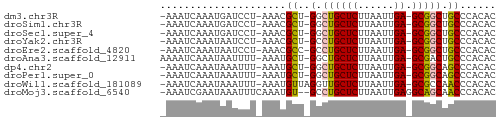
>dm3.chr3R 25614379 52 - 27905053 -AAAUCAAAUGAUCCU-AAACGCU-GGCUGCUCUUAAUUGA-GCGGCUGCCCACAC -...............-....((.-((((((((......))-))))))))...... ( -15.60, z-score = -3.42, R) >droSim1.chr3R 25286175 52 - 27517382 -AAAUCAAAUGAUCCU-AAACGCU-GGCUGCUCUUAAUUGA-GCGGCUGCCCACAC -...............-....((.-((((((((......))-))))))))...... ( -15.60, z-score = -3.42, R) >droSec1.super_4 4465006 52 - 6179234 -AAAUCAAAUGAUCCU-AAACGCU-GGCUGCUCUUAAUUGA-GCGGCUGCCCACAC -...............-....((.-((((((((......))-))))))))...... ( -15.60, z-score = -3.42, R) >droYak2.chr3R 7977899 52 + 28832112 -AAAUCAAAUAAUCCU-AAACGCU-GCCUGCUCUUAAUUGA-GCGGCUGCCCACAC -...............-....((.-(((.((((......))-))))).))...... ( -13.90, z-score = -4.02, R) >droEre2.scaffold_4820 8081150 52 + 10470090 -AAAUCAAAUAAUCCU-AAACGCC-GCCUGCUCUUAAUUGA-GCGGCUGCCCACAC -...............-....((.-(((.((((......))-))))).))...... ( -13.90, z-score = -3.97, R) >droAna3.scaffold_12911 5317244 53 - 5364042 AAAAUCAAAUAAUUUU-AAAUGCU-GGCUGCUCUUAAUUGA-GCGACUGCCCACAC ................-...((..-((((((((......))-)))...)))..)). ( -10.10, z-score = -2.00, R) >dp4.chr2 24055733 52 + 30794189 -AAAUCAAAUAAAUUU-AAAUGCU-GGCUGCUCUUAAUUGA-GCGGCAGCCCACAC -...............-....(((-(.((((((......))-))))))))...... ( -16.00, z-score = -3.30, R) >droPer1.super_0 11248218 52 + 11822988 -AAAUCAAAUAAAUUU-AAAUGCU-GGCUGCUCUUAAUUGA-GCGGCAGCCCACAC -...............-....(((-(.((((((......))-))))))))...... ( -16.00, z-score = -3.30, R) >droWil1.scaffold_181089 5907341 53 - 12369635 -AAAUCAAAUAAAUUU-AAAUGUUAGGUUGCUCUUAAUUGA-GCGCCAACCCACAC -...............-....(((.(((.((((......))-))))))))...... ( -10.10, z-score = -2.38, R) >droMoj3.scaffold_6540 26940000 53 - 34148556 -AAAUCGAAUAAAUUUCAAAUGU--GCCUGCUCUUAAUUGAGGCAGCAACCCACAC -.....(((.....)))...(((--(.((((.(((....))))))).....)))). ( -8.90, z-score = -1.24, R) >consensus _AAAUCAAAUAAUCCU_AAACGCU_GGCUGCUCUUAAUUGA_GCGGCUGCCCACAC .....................((....((((((......)).))))..))...... ( -5.27 = -5.06 + -0.21)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:55:35 2011