
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 25,607,892 – 25,607,993 |
| Length | 101 |
| Max. P | 0.614207 |

| Location | 25,607,892 – 25,607,993 |
|---|---|
| Length | 101 |
| Sequences | 7 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 76.10 |
| Shannon entropy | 0.46562 |
| G+C content | 0.42005 |
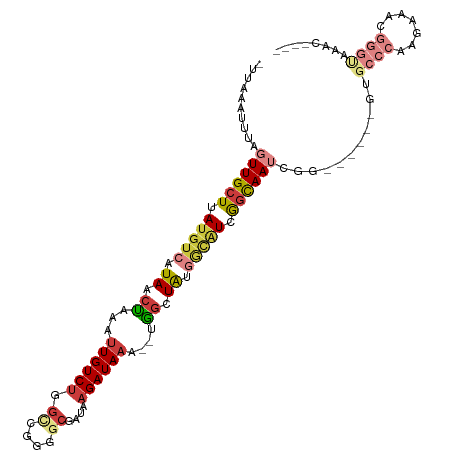
| Mean single sequence MFE | -25.79 |
| Consensus MFE | -12.11 |
| Energy contribution | -14.07 |
| Covariance contribution | 1.96 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.614207 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
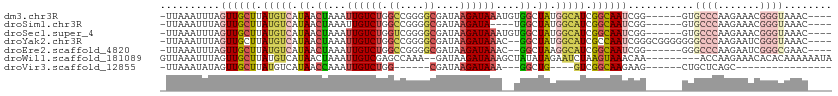
>dm3.chr3R 25607892 101 + 27905053 -UUAAAUUUAGUUGCUUAUGUCAUAACUAAAUUGUCUGGCCGGGGCGAUAAGAUAAAUGUGGCUAUGGCAUCGGCAAUCGG------GUGCCCAAGAAACGGGUAAAC---- -....(((..((((((.((((((((.(((..((((((.((....))....))))))...))).)))))))).))))))..)------))((((.......))))....---- ( -32.90, z-score = -3.09, R) >droSim1.chr3R 25279869 97 + 27517382 -UUAAAUUUAGUUGCUUAUGUCAUAACUAAAUUGUCUGGCCGGGGCGAUAAGAUA----UGGCUAUGGCAUCGGCAAUCGG------GUGCCCAAGAAACGGGUAAAC---- -....(((..((((((.((((((((.(((...(((((.((....))....)))))----))).)))))))).))))))..)------))((((.......))))....---- ( -30.40, z-score = -2.49, R) >droSec1.super_4 4458698 101 + 6179234 -UUAAAUUUAGUUGCUUAUGUCAUAACUAAAUUGUCUGGUCGGGGCGAUAAGAUAAAUGUGGCUAUGGCAUCGGCAAUCGG------GUGCCCAAGAAACGGGUAAAC---- -....(((..((((((.((((((((.(((..((((((.((((...)))).))))))...))).)))))))).))))))..)------))((((.......))))....---- ( -33.90, z-score = -4.10, R) >droYak2.chr3R 7970588 105 - 28832112 -UUAAAUUUAGUUGCUUAUGUCAUAACUAAAUUGUCUGGCCGGGGCGAUAAGAUAAAC--GGCUAUGGCAUCGCCAAUCGGGCGGGGGGGCCCAAGAAUCGGGUAAAC---- -...((((((((((.........)))))))))).((((.(((.((((((..(.....)--.((....))))))))...))).))))...((((.......))))....---- ( -29.70, z-score = -0.75, R) >droEre2.scaffold_4820 8074753 99 - 10470090 -UUAAAUUUAGUUGCUUAUGUCAUAACUAAAUUGUCUGGCCGGGGCGAUAAGAUAAAC--GGCUAAGGCAUCGGCAAUCGG------GGGCCCAAGAAUCGGGCGAAC---- -.....((..((((((.(((((...((......)).((((((...(.....).....)--))))).))))).))))))..)------).((((.......))))....---- ( -28.40, z-score = -1.46, R) >droWil1.scaffold_181089 5898147 101 + 12369635 GUUAAAUUUAGUUGCUUAUGUCAUAACUAAAUUGUCGAGCCAAA--GAUAAGAUAAAGCUAUAUAGAAUCUAAGUAAACAA---------ACCAAGAAACACACAAAAAAUA (((...((((...((((.((((.........(((((........--)))))))))))))....)))).(((..((......---------))..))))))............ ( -8.70, z-score = 0.33, R) >droVir3.scaffold_12855 1179150 76 + 10161210 -UUAAAUAUAGUUGCUUAUGUCAUAACCAAAUUGUCUGG------CGAUAAGAUAAA---GGCUG----GUCGGCAAGAAG------CUGCUCAGC---------------- -.........((((((...(.((.........)).).))------))))........---.((((----..((((.....)------)))..))))---------------- ( -16.50, z-score = -0.29, R) >consensus _UUAAAUUUAGUUGCUUAUGUCAUAACUAAAUUGUCUGGCCGGGGCGAUAAGAUAAA__UGGCUAUGGCAUCGGCAAUCGG______GUGCCCAAGAAACGGGUAAAC____ ..........((((((.(((((.((.((...((((((.((....))....))))))....)).)).))))).))))))...........((((.......))))........ (-12.11 = -14.07 + 1.96)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:55:34 2011