| Sequence ID | dm3.chr2L |
|---|---|
| Location | 9,125,810 – 9,125,900 |
| Length | 90 |
| Max. P | 0.559222 |

| Location | 9,125,810 – 9,125,900 |
|---|---|
| Length | 90 |
| Sequences | 8 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 77.77 |
| Shannon entropy | 0.41371 |
| G+C content | 0.34885 |
| Mean single sequence MFE | -15.33 |
| Consensus MFE | -11.02 |
| Energy contribution | -11.15 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.96 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.559222 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 9125810 90 + 23011544 CACUUUGAAAAUAAAAUUUUGGCCAA-UAACAAAAUUGGUUUUUUCAAUUUGCUAAGCCAC---AGCCGCA--------CCGAGAAACCGCAAAGAAAUGCG- ..(((((.......(((((((.....-...)))))))((((((((.....(((...((...---.)).)))--------..)))))))).))))).......- ( -17.10, z-score = -1.09, R) >droSim1.chr2L 8905910 90 + 22036055 CAAUUUGAAAAUAAAAUUUUGGCCAA-UAACAAAAUUGGUUUUUUCAAUUUGCUAAGCCAC---AGCCGCA--------CCGAGAAACCGCAAAGAAAUGCG- ..............(((((((.....-...)))))))((((((((.....(((...((...---.)).)))--------..))))))))(((......))).- ( -15.20, z-score = -0.39, R) >droSec1.super_3 4591907 90 + 7220098 CAAUUUGAAAAUAAAAUUUUGGCCAA-UAACAAAAUUGGUUUUUUCAAUUUGCUAAGCCAC---AGCCGCA--------CCGAGAAACCGCAAAGAAAUGCG- ..............(((((((.....-...)))))))((((((((.....(((...((...---.)).)))--------..))))))))(((......))).- ( -15.20, z-score = -0.39, R) >droYak2.chr2L 11792333 101 + 22324452 CAAUUUGAAAAUAAAAUUUUGGCCAA-UAACAAAAUUGGUUUUUUCAAUUUGCCAAGCGACAACAGCCGCAUCGAGAAACCGAGAAACCGCAAAGAAAUGCG- (((.(((((((...(((((((.....-...)))))))....))))))).)))....(((........))).(((......))).....((((......))))- ( -19.80, z-score = -1.30, R) >droEre2.scaffold_4929 9730265 90 + 26641161 CAAUUUGAAAAUAAAAUUUUGGCCAA-UAACAAAAUUGGUUUUUUCAAUU-GCCAAGCCAUU--AGCCGCA--------CCGAGAAACCGCAAAGAAAUGCG- ..............(((((((.....-...)))))))((((((((....(-((...((....--.)).)))--------..))))))))(((......))).- ( -15.90, z-score = -0.65, R) >droAna3.scaffold_12943 474186 94 + 5039921 CGAUUUGAAAAUAAAAUUUUGCCCAA-AAACAAAAUUGGUUUUUUCAAUU--GCAAAUCAUUACAUGAGAACC-----AAGAAGAAACCGCAAAGAAAUACG- ...((((.......(((((((.....-...)))))))(((((((((..((--(....((((...))))....)-----))))))))))).))))........- ( -15.40, z-score = -2.24, R) >dp4.chr4_group5 1432431 72 + 2436548 GAAUUUGAAAAUAAAAUUUUGCCCCGCUAACAAAAUUGGUUUUUUCAAUUG-------------------------------CCAAAACGCAAAGAAAUGCGA ....(((((((...(((((((.........)))))))....)))))))...-------------------------------......((((......)))). ( -12.00, z-score = -0.82, R) >droPer1.super_5 5839072 72 + 6813705 GAAUUUGAAAAUAAAAUUUUGCCCCGCUAACAAAAUUGGUUUUUUCAAUUG-------------------------------CCAAAACGCAAAGAAAUGCGA ....(((((((...(((((((.........)))))))....)))))))...-------------------------------......((((......)))). ( -12.00, z-score = -0.82, R) >consensus CAAUUUGAAAAUAAAAUUUUGGCCAA_UAACAAAAUUGGUUUUUUCAAUUUGCCAAGCCAC___AGCCGCA________CCGAGAAACCGCAAAGAAAUGCG_ ....(((((((...(((((((.........)))))))....)))))))........................................((((......)))). (-11.02 = -11.15 + 0.13)


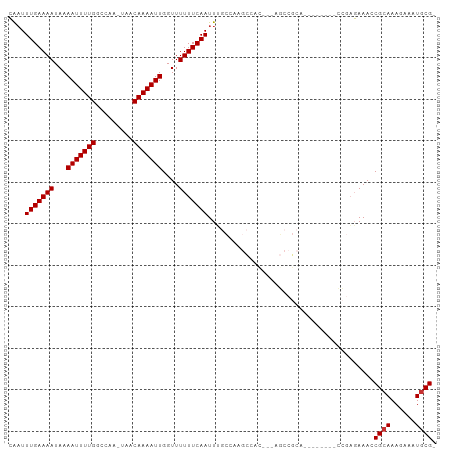
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:27:37 2011