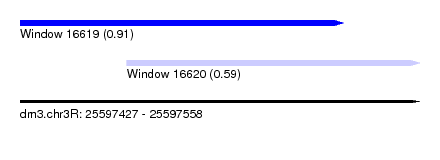
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 25,597,427 – 25,597,558 |
| Length | 131 |
| Max. P | 0.911815 |

| Location | 25,597,427 – 25,597,533 |
|---|---|
| Length | 106 |
| Sequences | 7 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 78.61 |
| Shannon entropy | 0.42321 |
| G+C content | 0.50777 |
| Mean single sequence MFE | -34.33 |
| Consensus MFE | -19.67 |
| Energy contribution | -19.86 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.911815 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
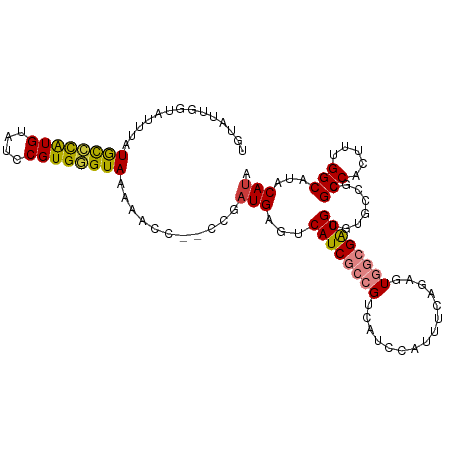
>dm3.chr3R 25597427 106 + 27905053 UGUGUUGGUAUUUAUGCCCAUGUAUCCGUGGGUAAAAACC--CCGAUGAGUCAUCGCCGUCAUCCAUUUCAGAGUGGCGAUGCGUGCCGCGCCACUUUGGCAUACAUA (((((.((..(((.((((((((....)))))))).)))..--))((((.((....))))))........((((((((((..(....)..))))))))))))))).... ( -37.30, z-score = -2.27, R) >droSim1.chr3R 25269532 106 + 27517382 UGUGUUGGUAUUUAUGCCCAUGUAUCCGUGGGUAAAAACC--CCGAUGAGUCAUCGCCGUCAUCCAUUUCAGAGUGGCGAUGCGUGCCGCGCCACUUUGGCAUACAUA (((((.((..(((.((((((((....)))))))).)))..--))((((.((....))))))........((((((((((..(....)..))))))))))))))).... ( -37.30, z-score = -2.27, R) >droYak2.chr3R 7959015 105 - 28832112 UGUAUUGGUAUUUAUGCCCAUGUAUCCGUGGGUAAAACCC---CGAUGAGUCAUCGCCGUCAUCCAUUUCAGAGUGGCGAUGCGUGCCGCGCCACUUUGGCAUACAUA ..((((((..(((.((((((((....)))))))))))..)---))))).(.((((((((((..........)).)))))))))((((((........))))))..... ( -36.60, z-score = -2.25, R) >droEre2.scaffold_4820 8064035 105 - 10470090 UGUAUUGGUAUUUAUGCCCAUGUACCCGUGGGUAAAACCC---CGAUGACUCAUCGCCGUCAUCCAUUUCAGAGUGGCGAUGCGUGCCGCGCCACUUUGGCAUACAUA (((((.((..(((.((((((((....)))))))))))..)---)((((((........))))))....(((((((((((..(....)..))))))))))).))))).. ( -39.50, z-score = -3.19, R) >droAna3.scaffold_12911 5298786 92 + 5364042 UGUAUUUGUAUCUGUAUCCGUGU--CCGUGUGUAAAAACC--CCGAUGAGUCAUCA--GGCA----------AAUGCCGAUGCGUGCUGCGCCACAUUGGCAUACAUA ......(((((..(((.((((((--(..............--..((((...)))).--(((.----------...))))))))).).)))(((.....)))))))).. ( -22.40, z-score = 0.25, R) >droVir3.scaffold_12855 1165307 93 + 10161210 --------UGUGUGUGUGUGUGUGACCGUGUGUAGAAACC--CCGAUGAGUCAUCA--GCGGUCCAAGC--GAGUGA-GGUGCGUGGGUCGCCACAUUGGCAUACAAA --------((((((((((((.((((((.............--(((.(((....)))--.)))..((.((--(.....-..))).)))))))))))))..))))))).. ( -31.30, z-score = -1.04, R) >droSec1.super_4 4448546 98 + 6179234 ----------UUUAUGCCCAUGUAUCCGUGGGUAAAAGCUGUUUGAUGAGUCAUCGCCGUCAUCCAUUUCAGAGUGGCGAUGCUUGCCGCGCCACUUUGGCAUACAUA ----------....((((((((....))))))))...((((..(((((.((....)))))))..))...((((((((((..(....)..))))))))))))....... ( -35.90, z-score = -3.12, R) >consensus UGUAUUGGUAUUUAUGCCCAUGUAUCCGUGGGUAAAAACC__CCGAUGAGUCAUCGCCGUCAUCCAUUUCAGAGUGGCGAUGCGUGCCGCGCCACUUUGGCAUACAUA ..............((((((((....))))))))..........((((...))))..(((((..(......)..)))))(((..(((((........)))))..))). (-19.67 = -19.86 + 0.19)

| Location | 25,597,462 – 25,597,558 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 93.69 |
| Shannon entropy | 0.11230 |
| G+C content | 0.49383 |
| Mean single sequence MFE | -25.86 |
| Consensus MFE | -22.24 |
| Energy contribution | -22.44 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.593600 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
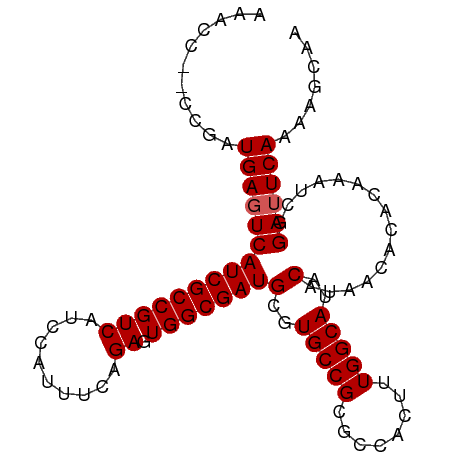
>dm3.chr3R 25597462 96 + 27905053 AAACC--CCGAUGAGUCAUCGCCGUCAUCCAUUUCAGAGUGGCGAUGCGUGCCGCGCCACUUUGGCAUACAUAACACACAAAUCGGAUUCAAAAGCAA .....--(((((..(.((((((((((..........)).)))))))))((((((........)))))).............)))))............ ( -25.80, z-score = -1.41, R) >droSim1.chr3R 25269567 96 + 27517382 AAACC--CCGAUGAGUCAUCGCCGUCAUCCAUUUCAGAGUGGCGAUGCGUGCCGCGCCACUUUGGCAUACAUAACACACAAAUCGGAUUCAAAAGCAA .....--(((((..(.((((((((((..........)).)))))))))((((((........)))))).............)))))............ ( -25.80, z-score = -1.41, R) >droYak2.chr3R 7959050 95 - 28832112 -AACC--CCGAUGAGUCAUCGCCGUCAUCCAUUUCAGAGUGGCGAUGCGUGCCGCGCCACUUUGGCAUACAUAACACACAAAUCGGAUUCAAAACCAA -....--(((((..(.((((((((((..........)).)))))))))((((((........)))))).............)))))............ ( -25.80, z-score = -1.74, R) >droEre2.scaffold_4820 8064070 91 - 10470090 -AACC--CCGAUGACUCAUCGCCGUCAUCCAUUUCAGAGUGGCGAUGCGUGCCGCGCCACUUUGGCAUACAUAACACACAAAUCUGAUUCAAAA---- -....--..((((((........))))))....(((((((((((.....))))))(((.....)))................))))).......---- ( -25.90, z-score = -2.67, R) >droSec1.super_4 4448571 98 + 6179234 AAGCUGUUUGAUGAGUCAUCGCCGUCAUCCAUUUCAGAGUGGCGAUGCUUGCCGCGCCACUUUGGCAUACAUAACACACAAAUCGGAUUCAAAAGCAA ..((((..(((((.((....)))))))..)...(((((((((((..(....)..)))))))))))............................))).. ( -26.00, z-score = -0.82, R) >consensus AAACC__CCGAUGAGUCAUCGCCGUCAUCCAUUUCAGAGUGGCGAUGCGUGCCGCGCCACUUUGGCAUACAUAACACACAAAUCGGAUUCAAAAGCAA ...........(((((((((((((((..........)).)))))))(..(((((........)))))..)...............))))))....... (-22.24 = -22.44 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:55:30 2011