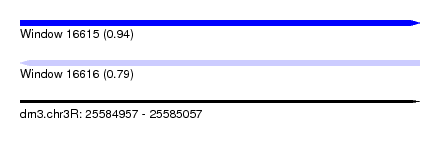
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 25,584,957 – 25,585,057 |
| Length | 100 |
| Max. P | 0.938799 |

| Location | 25,584,957 – 25,585,057 |
|---|---|
| Length | 100 |
| Sequences | 9 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 72.84 |
| Shannon entropy | 0.57091 |
| G+C content | 0.52970 |
| Mean single sequence MFE | -25.76 |
| Consensus MFE | -14.89 |
| Energy contribution | -14.96 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.938799 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
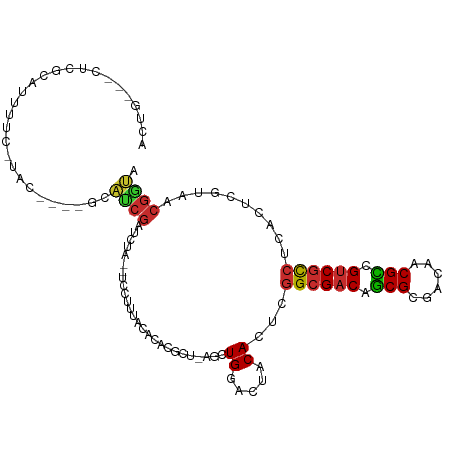
>dm3.chr3R 25584957 100 + 27905053 ACUG---AUCGUAUUUUC-UAC----GCAUCGAUCUA--UCCUUUACACACGCU-AGCUGGACUACACUAGGCGACAGCGCGACAACGCCGUCGCCUCACUCGUAACGGUA ..((---((((((.....-)))----).)))).....--.((.((((......(-((.((.....)))))((((((.(((......))).))))))......)))).)).. ( -26.50, z-score = -1.57, R) >droSim1.chr2h_random 1859885 100 + 3178526 ACUG---AUCGUAUUUUC-UAC----GCAUCGAUCUA--UCCUUUACACACCCU-AGCUGGACUACACUUGGCGACAGCGAGACAACGCCGUCGCCUCACUCGUAACGGUA ..((---((((((.....-)))----).)))).....--.((.((((....((.-....)).........((((((.(((......))).))))))......)))).)).. ( -26.30, z-score = -2.22, R) >droSec1.super_18 1176912 100 + 1342155 ACUG---AUCGUAUUUUC-UAC----GCAUCGAUCUA--UCCUUUACACACGCU-AGCUGGACUACACUUGGCGACAGCGCGACAACGCCGUCGCCUCACUCGUAACGGUA ..((---((((((.....-)))----).)))).....--.((.((((...((((-((.((.....)).))))))..((.(((((......))))))).....)))).)).. ( -25.50, z-score = -1.34, R) >droEre2.scaffold_4690 11531505 104 - 18748788 ACUG---CUCGCAUUUUC-GACUCGCGCACCGAUCUA--UGCUUUACACACGCU-AACUGGACUACACAUGGCGACAGCGCGACAACGCCGUCGCCUCACACGUAACGGUA ....---...((((..((-(..........)))...)--)))............-.((((..........((((((.(((......))).))))))..........)))). ( -25.15, z-score = -0.12, R) >droYak2.chr4 88941 100 - 1374474 ACUG---CUCGCAUUUUC-UAC----GCAUCGAUCUA--UCCUUUACACACGCU-AGCUGGACUACACUAGGCGACAACGCGACAACGUCGUCGCCCCACUCGUUGCGGUA ....---...........-..(----(((.(((....--..............(-((.((.....)))))((((((.(((......))).))))))....))).))))... ( -24.10, z-score = -0.72, R) >droAna3.scaffold_13339 844766 96 - 4599533 --------UGGCACUGCC-------UGCGUCGUCCCUUGUUCUUUCUAUCCUUAUCACUGAACCACACUCGGCGACAGCGACACUACGCUGUCGCCCUGCUAAAAGCGACA --------.(((...)))-------...(((((.....((((.................)))).......((((((((((......)))))))))).........))))). ( -30.73, z-score = -3.77, R) >droWil1.scaffold_180875 4102 106 - 26602 ACUGA--CUCGCAUUCUU-CGAUCACGCAUCGAUCUAC-UUCUUUUUAGACGCCAAGCUGGACUACACUCGGCGACAGCGCGACAACGCCGUCGCCUCACUCAUA-CGGUA ((((.--((.((.....(-((((.....)))))((((.-.......))))......)).)).........((((((.(((......))).)))))).........-)))). ( -26.30, z-score = -1.24, R) >droVir3.scaffold_12958 3169031 101 + 3547706 UAUG---CUGGCAUUUUC-UACUCACUCAUCGAUCUA--UGUGUAACACGCA---AGCUGGACUACACUCGGCGUCAGCGCGACAACGCCGUCCCCUCACUCGUA-CGGUA ...(---(((((......-..................--((((.....))))---.(((((.......)))))))))))........(((((.(........).)-)))). ( -23.60, z-score = 0.07, R) >droMoj3.scaffold_6498 896728 103 - 3408170 ACUGAUCUUCUUAUUCUCGCAUCAGUACCUGCUUCUGU-UGCUUUUCAAACGCA-AGUUGGACUACACUCAGCGACAGCGCGACACCGUCGUCGUCUCGCUCAAA------ (((((((...........).)))))).((.((((.(((-(........)))).)-))).)).........(((((..(((((((...)))).))).)))))....------ ( -23.70, z-score = -0.79, R) >consensus ACUG___CUCGCAUUUUC_UAC____GCAUCGAUCUA__UCCUUUACACACGCU_AGCUGGACUACACUCGGCGACAGCGCGACAACGCCGUCGCCUCACUCGUAACGGUA ............................((((..........................((.....))...((((((.(((......))).))))))..........)))). (-14.89 = -14.96 + 0.06)

| Location | 25,584,957 – 25,585,057 |
|---|---|
| Length | 100 |
| Sequences | 9 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 72.84 |
| Shannon entropy | 0.57091 |
| G+C content | 0.52970 |
| Mean single sequence MFE | -33.11 |
| Consensus MFE | -18.41 |
| Energy contribution | -18.49 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.00 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.790012 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
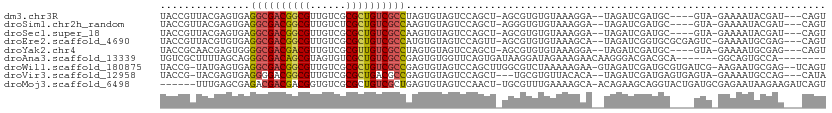
>dm3.chr3R 25584957 100 - 27905053 UACCGUUACGAGUGAGGCGACGGCGUUGUCGCGCUGUCGCCUAGUGUAGUCCAGCU-AGCGUGUGUAAAGGA--UAGAUCGAUGC----GUA-GAAAAUACGAU---CAGU ..((.(((((.((.((((((((((((....))))))))))))....(((.....))-)))...))))).)).--......(((.(----(((-.....))))))---)... ( -35.50, z-score = -1.91, R) >droSim1.chr2h_random 1859885 100 - 3178526 UACCGUUACGAGUGAGGCGACGGCGUUGUCUCGCUGUCGCCAAGUGUAGUCCAGCU-AGGGUGUGUAAAGGA--UAGAUCGAUGC----GUA-GAAAAUACGAU---CAGU ..((.(((((.....((((((((((......)))))))))).(.(.(((.....))-).).).))))).)).--......(((.(----(((-.....))))))---)... ( -33.30, z-score = -1.85, R) >droSec1.super_18 1176912 100 - 1342155 UACCGUUACGAGUGAGGCGACGGCGUUGUCGCGCUGUCGCCAAGUGUAGUCCAGCU-AGCGUGUGUAAAGGA--UAGAUCGAUGC----GUA-GAAAAUACGAU---CAGU ..((.(((((.((..(((((((((((....))))))))))).....(((.....))-)))...))))).)).--......(((.(----(((-.....))))))---)... ( -34.70, z-score = -1.90, R) >droEre2.scaffold_4690 11531505 104 + 18748788 UACCGUUACGUGUGAGGCGACGGCGUUGUCGCGCUGUCGCCAUGUGUAGUCCAGUU-AGCGUGUGUAAAGCA--UAGAUCGGUGCGCGAGUC-GAAAAUGCGAG---CAGU ....(((.(((((..(((((((((((....))))))))))).((((((..((.(((-...((((.....)))--).))).))))))))....-....)))))))---)... ( -35.60, z-score = -0.15, R) >droYak2.chr4 88941 100 + 1374474 UACCGCAACGAGUGGGGCGACGACGUUGUCGCGUUGUCGCCUAGUGUAGUCCAGCU-AGCGUGUGUAAAGGA--UAGAUCGAUGC----GUA-GAAAAUGCGAG---CAGU ..((((.....))))(((((((((((....)))))))))))............(((-.((((.......(.(--(......)).)----...-....)))).))---)... ( -32.54, z-score = -0.65, R) >droAna3.scaffold_13339 844766 96 + 4599533 UGUCGCUUUUAGCAGGGCGACAGCGUAGUGUCGCUGUCGCCGAGUGUGGUUCAGUGAUAAGGAUAGAAAGAACAAGGGACGACGCA-------GGCAGUGCCA-------- ((((((.....))..((((((((((......))))))))))..((((.((((..((................))..)))).)))).-------))))......-------- ( -36.69, z-score = -2.67, R) >droWil1.scaffold_180875 4102 106 + 26602 UACCG-UAUGAGUGAGGCGACGGCGUUGUCGCGCUGUCGCCGAGUGUAGUCCAGCUUGGCGUCUAAAAAGAA-GUAGAUCGAUGCGUGAUCG-AAGAAUGCGAG--UCAGU .((((-(((.....((((((((((((....))))))))(((((((........)))))))))))........-.....(((((.....))))-)...))))).)--).... ( -36.60, z-score = -1.51, R) >droVir3.scaffold_12958 3169031 101 - 3547706 UACCG-UACGAGUGAGGGGACGGCGUUGUCGCGCUGACGCCGAGUGUAGUCCAGCU---UGCGUGUUACACA--UAGAUCGAUGAGUGAGUA-GAAAAUGCCAG---CAUA .((..-(((......(....)((((((........))))))..)))..))...(((---.((((.((((.((--(..........))).)))-)...)))).))---)... ( -26.20, z-score = 1.22, R) >droMoj3.scaffold_6498 896728 103 + 3408170 ------UUUGAGCGAGACGACGACGGUGUCGCGCUGUCGCUGAGUGUAGUCCAACU-UGCGUUUGAAAAGCA-ACAGAAGCAGGUACUGAUGCGAGAAUAAGAAGAUCAGU ------.((.(((((.(((.((((...)))))).).))))).)).........(((-(((.((((.......-.)))).))))))((((((.(........)...)))))) ( -26.90, z-score = 0.45, R) >consensus UACCGUUACGAGUGAGGCGACGGCGUUGUCGCGCUGUCGCCGAGUGUAGUCCAGCU_AGCGUGUGUAAAGGA__UAGAUCGAUGC____GUA_GAAAAUGCGAG___CAGU ...............((((((((((......))))))))))...................................................................... (-18.41 = -18.49 + 0.08)
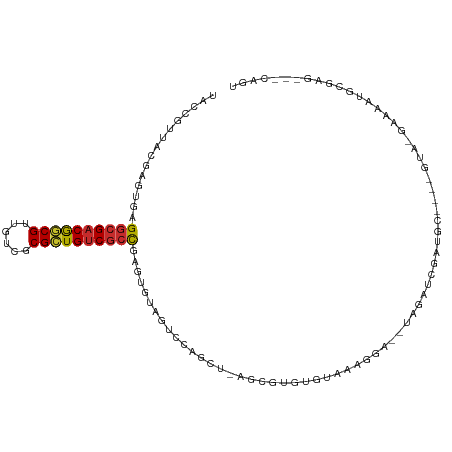
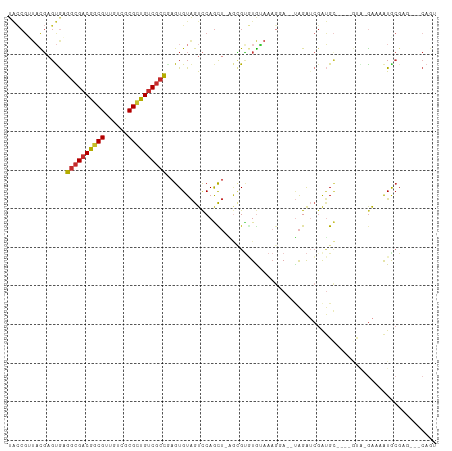
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:55:27 2011