
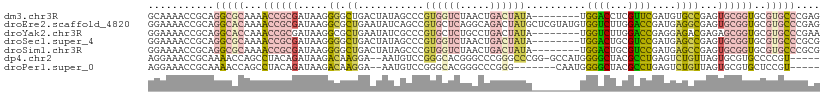
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 25,583,245 – 25,583,349 |
| Length | 104 |
| Max. P | 0.911828 |

| Location | 25,583,245 – 25,583,349 |
|---|---|
| Length | 104 |
| Sequences | 7 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 63.40 |
| Shannon entropy | 0.68326 |
| G+C content | 0.59859 |
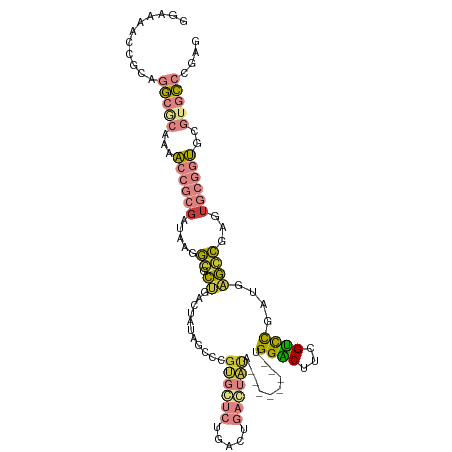
| Mean single sequence MFE | -40.29 |
| Consensus MFE | -16.05 |
| Energy contribution | -16.23 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.56 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.911828 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 25583245 104 + 27905053 GCAAAACCGCAGGCGCAAAACCGCGAUAAGGGGCUGACUAUAGCCCGUGGUCUAACUGACUAUA--------UGGACCUCGUUCGAUGUGCCGAGUGCGGUGCGUGCCCGAG (((..((((((((((((.....((((....((((((....))))))((((((.....)))))).--------......))))....))))))...))))))...)))..... ( -40.80, z-score = -2.19, R) >droEre2.scaffold_4820 8051033 112 - 10470090 GGAAAACCGCAGGCACAAAACCGCGAUAAGGCGCUGAAUAUCAGCCGUGCUCAGGCAGACUAUGCUCGUAUGUGGUCUUGGACCGAUGAGGCGAGUGCGGUGCGUGCCCGAG ((....))...(((((...(((((....(((.((((.....))))).(((....)))..))..((((((...(((((...))))).....)))))))))))..))))).... ( -39.70, z-score = -0.46, R) >droYak2.chr3R 7945594 104 - 28832112 GGAAAACCGCAGGCACCAAACCGCGAUAAGGCGCUGAAUAUCGCCCGUGCUCUGCCUGACUAUA--------UGGUCUUGGACCGAGGAGACGAGAGCGGUGCGUGCCCGAA ((....))(((.(((((.....(((....((((........)))))))(((((.((.((((...--------.))))..)).....(....).)))))))))).)))..... ( -36.70, z-score = -1.32, R) >droSec1.super_4 4435970 104 + 6179234 GGAAAACCGCAGGCGCAAAACCGCGAUAAGGGGCUGACUAUAGCCCGUGGUCUAACUGACUAUA--------UGGACUGCGUCCGAUGAGCCGAGUGCGGUGCGUGCCCGCG .......(((.(((((...((((((.....((((((....))))))((((((.....)))))).--------(((((...)))))..........))))))..))))).))) ( -41.80, z-score = -2.08, R) >droSim1.chr3R 25256313 104 + 27517382 GGAAAACCGCAGGCGCAAAACCGCGAUAAGGGGCUGACUAUAGCCCGUGGUCUAACUGACUAUA--------UGGACUGCGUCCGAUGAGCCGAGUGCGGUGCGUGCCCGCG .......(((.(((((...((((((.....((((((....))))))((((((.....)))))).--------(((((...)))))..........))))))..))))).))) ( -41.80, z-score = -2.08, R) >dp4.chr2 24022570 104 - 30794189 AGGAAACCGCAAAACCAGCCUACAGAUAAGACAAGGA--AAUGUCCGGGCACGGGCCCGGGCCCGG-GCCAUGGGGCUACGCCUGAGUCUGUUAGUGCGUGCCCCGU----- .((...(((.........(((............))).--...((((((((....)))))))).)))-.))(((((((.((((((((.....)))).)))))))))))----- ( -44.00, z-score = -1.21, R) >droPer1.super_0 11214151 98 - 11822988 AGGAAACCGCAAAACCAGCCUACAGAUAAGACAAGGA--AAUGUCCGGGCACGGGCCCGGG-------CAAUGGGGCUACGCCUGAGUCUGUUAGUGCGUGCUCCGU----- .(....)...........(((............))).--..(((((((((....)))))))-------))(((((((.((((((((.....)))).)))))))))))----- ( -37.20, z-score = -1.30, R) >consensus GGAAAACCGCAGGCGCAAAACCGCGAUAAGGCGCUGACUAUAGCCCGUGCUCUGACUGACUAUA________UGGACUUCGUCCGAUGAGCCGAGUGCGGUGCGUGCCCGAG ...........(((((...((((((.....((.((...........((((((.....))))))..........((((...))))....))))...))))))..))))).... (-16.05 = -16.23 + 0.17)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:55:25 2011