| Sequence ID | dm3.chr2L |
|---|---|
| Location | 9,122,692 – 9,122,788 |
| Length | 96 |
| Max. P | 0.722871 |

| Location | 9,122,692 – 9,122,788 |
|---|---|
| Length | 96 |
| Sequences | 7 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 80.38 |
| Shannon entropy | 0.38916 |
| G+C content | 0.37505 |
| Mean single sequence MFE | -23.58 |
| Consensus MFE | -11.87 |
| Energy contribution | -12.01 |
| Covariance contribution | 0.15 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.722871 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
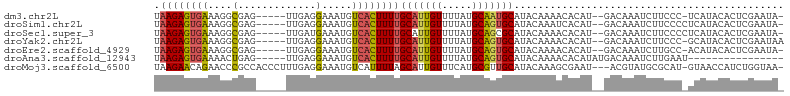
>dm3.chr2L 9122692 96 + 23011544 UAAGAGUGAAAGGCGAG-----UUGAGGAAAUGUCACUUUUGCAUUGUUUUAUGCAAUGCAUACAAAACACAU--GACAAAUCUUCCC-UCAUACACUCGAAUA- ...(((((......(((-----..(((((..(((((....((((((((.....))))))))...........)--))))..))))).)-))...))))).....- ( -25.36, z-score = -3.00, R) >droSim1.chr2L 8902776 97 + 22036055 UAAGAGUGAAAGGCGAG-----UUGAGGAAAUGUCACUUUUGCAUUGUUUUAUGCAGUGCAUACAAAUCACAU--GACAAAUCUUCCCCUCAUACACUCGAAUA- ...(((((......(((-----..(((((..(((((....((((((((.....))))))))...........)--))))..)))))..)))...))))).....- ( -25.86, z-score = -2.60, R) >droSec1.super_3 4588796 97 + 7220098 UAAGAGUGAAAGGCGAG-----UUGAUGAAAUGUCACUUUUGCAUUGUUUUAUGCAGCGCAUACAAAACACAU--GACAAAUCUUCCCCUCAUACACUCGAAUA- ...(((((..(((.(((-----.((((.....))))..(((((((((((((((((...)))))..))))).))--).))))..))).)))....))))).....- ( -20.30, z-score = -1.21, R) >droYak2.chr2L 11789062 97 + 22324452 UAAGAGUGAAAGGCGAG-----UUGAGGAAAUGUCACUUUUGCAUUGUUUUAUGCAGUGCAUACAAAACACAU--GACAAAUCUUCCC-GCAUACACUCGAAUAA ...(((((....(((.(-----..(((((..(((((....((((((((.....))))))))...........)--))))..)))))))-))...)))))...... ( -27.86, z-score = -3.29, R) >droEre2.scaffold_4929 9727142 96 + 26641161 UAAGAGUGAAAGGCGAG-----UUGAGGAAAUGUCACUUUUGCAUUGUUUUAUGCAGUGCAUACAAAACACAU--GACAAAUCUUGCC-ACAUACACUCGAAUA- ...(((((...((((((-----.....(..((((......((((((((.....))))))))........))))--..)....))))))-.....))))).....- ( -26.94, z-score = -2.86, R) >droAna3.scaffold_12943 470944 84 + 5039921 UAAGAGUGAAAACUGAG-----UUGAGGAAAUGUCACUUUUGCAUUGUUUUAUGCAGUGCAUACAAAACACAUAUGACAAAUCUUGAAU---------------- ....(((....)))..(-----((.((((..(((((....((((((((.....)))))))).............)))))..)))).)))---------------- ( -18.43, z-score = -1.41, R) >droMoj3.scaffold_6500 11974442 100 + 32352404 UAAGAACAGAACCCGCCACCCUUUGAGGAAAUGUCAUUUUAGCAUUGUUUCAUGCGUUGCAUACAAAGCGAAU---ACGUAUGCGCAU-GUAACCAUCUGGUAA- ......((((.............(((((.(((((.......))))).)))))((((((((((((.........---..))))))).))-)))....))))....- ( -20.30, z-score = -0.09, R) >consensus UAAGAGUGAAAGGCGAG_____UUGAGGAAAUGUCACUUUUGCAUUGUUUUAUGCAGUGCAUACAAAACACAU__GACAAAUCUUCCC_UCAUACACUCGAAUA_ .((((((((....(((......)))........))))))))(((((((.....)))))))............................................. (-11.87 = -12.01 + 0.15)
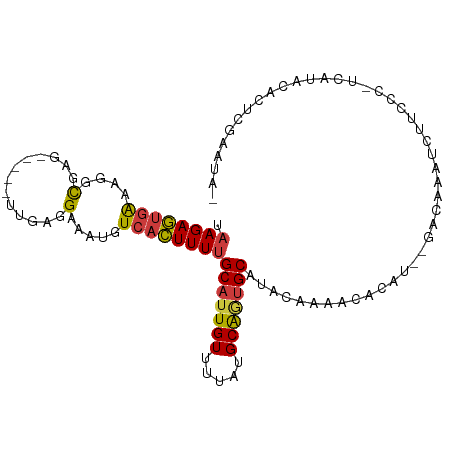
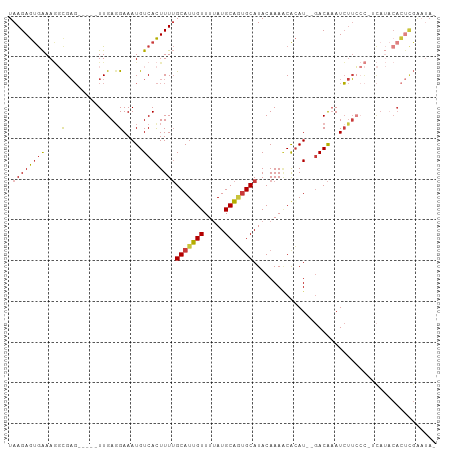
| Location | 9,122,692 – 9,122,788 |
|---|---|
| Length | 96 |
| Sequences | 7 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 80.38 |
| Shannon entropy | 0.38916 |
| G+C content | 0.37505 |
| Mean single sequence MFE | -25.39 |
| Consensus MFE | -9.92 |
| Energy contribution | -11.31 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.534820 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 9122692 96 - 23011544 -UAUUCGAGUGUAUGA-GGGAAGAUUUGUC--AUGUGUUUUGUAUGCAUUGCAUAAAACAAUGCAAAAGUGACAUUUCCUCAA-----CUCGCCUUUCACUCUUA -....(((((...(((-(((((....((((--((.(((.((((((((...))))...)))).)))...)))))))))))))))-----))))............. ( -27.50, z-score = -3.13, R) >droSim1.chr2L 8902776 97 - 22036055 -UAUUCGAGUGUAUGAGGGGAAGAUUUGUC--AUGUGAUUUGUAUGCACUGCAUAAAACAAUGCAAAAGUGACAUUUCCUCAA-----CUCGCCUUUCACUCUUA -.....(((((..((((..((.((..((((--((.((..((((((((...))))...))))..))...))))))..)).))..-----)))).....)))))... ( -28.00, z-score = -2.87, R) >droSec1.super_3 4588796 97 - 7220098 -UAUUCGAGUGUAUGAGGGGAAGAUUUGUC--AUGUGUUUUGUAUGCGCUGCAUAAAACAAUGCAAAAGUGACAUUUCAUCAA-----CUCGCCUUUCACUCUUA -.....(((((..((((..((.((..((((--((.(((.((((((((...))))...)))).)))...))))))..)).))..-----)))).....)))))... ( -30.30, z-score = -3.41, R) >droYak2.chr2L 11789062 97 - 22324452 UUAUUCGAGUGUAUGC-GGGAAGAUUUGUC--AUGUGUUUUGUAUGCACUGCAUAAAACAAUGCAAAAGUGACAUUUCCUCAA-----CUCGCCUUUCACUCUUA ......(((((...((-(((..((..((((--((((((.......))))(((((......)))))...))))))..)).....-----)))))....)))))... ( -29.00, z-score = -3.56, R) >droEre2.scaffold_4929 9727142 96 - 26641161 -UAUUCGAGUGUAUGU-GGCAAGAUUUGUC--AUGUGUUUUGUAUGCACUGCAUAAAACAAUGCAAAAGUGACAUUUCCUCAA-----CUCGCCUUUCACUCUUA -.....(((((.....-(((..((..((((--((((((.......))))(((((......)))))...))))))..)).....-----...)))...)))))... ( -24.60, z-score = -2.21, R) >droAna3.scaffold_12943 470944 84 - 5039921 ----------------AUUCAAGAUUUGUCAUAUGUGUUUUGUAUGCACUGCAUAAAACAAUGCAAAAGUGACAUUUCCUCAA-----CUCAGUUUUCACUCUUA ----------------......((..((((((..((((.......))))(((((......)))))...))))))..)).....-----................. ( -16.00, z-score = -1.43, R) >droMoj3.scaffold_6500 11974442 100 - 32352404 -UUACCAGAUGGUUAC-AUGCGCAUACGU---AUUCGCUUUGUAUGCAACGCAUGAAACAAUGCUAAAAUGACAUUUCCUCAAAGGGUGGCGGGUUCUGUUCUUA -....((((..(((.(-(((((((((((.---........)))))))...))))).)))..(((((...(((.......))).....)))))...))))...... ( -22.30, z-score = 0.11, R) >consensus _UAUUCGAGUGUAUGA_GGGAAGAUUUGUC__AUGUGUUUUGUAUGCACUGCAUAAAACAAUGCAAAAGUGACAUUUCCUCAA_____CUCGCCUUUCACUCUUA ......(((((........((.((..((((.....(((.((((((((...))))...)))).))).....))))..)).))................)))))... ( -9.92 = -11.31 + 1.39)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:27:36 2011