| Sequence ID | dm3.chr3R |
|---|---|
| Location | 25,537,169 – 25,537,275 |
| Length | 106 |
| Max. P | 0.964410 |

| Location | 25,537,169 – 25,537,275 |
|---|---|
| Length | 106 |
| Sequences | 9 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 72.21 |
| Shannon entropy | 0.57673 |
| G+C content | 0.59247 |
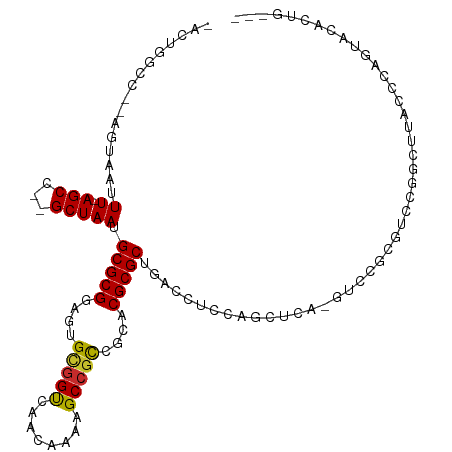
| Mean single sequence MFE | -34.98 |
| Consensus MFE | -16.60 |
| Energy contribution | -16.33 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.964410 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
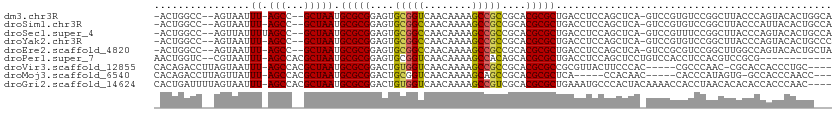
>dm3.chr3R 25537169 106 + 27905053 -ACUGGCC--AGUAAUUU-AGCC--GCUAAUGCGCGGAGUGCGGUCAACAAAAGCCGCCGCACGCGCUGACCUCCAGCUCA-GUCCGUGUCCGGCUUACCCAGUACACUGGCA -....(((--(((.....-..((--((......)))).(((((((......((((((..(((((.(((((.(....).)))-)).))))).)))))))))..)))))))))). ( -43.00, z-score = -2.42, R) >droSim1.chr3R 25209731 106 + 27517382 -ACUGGCC--AGUAAUUU-AGCC--GCUAAUGCGCGGAGUGCGGCCAACAAAAGCCGCCGCACGCGCUGACCUCCAGCUCA-GUCCGUGUCCGGCUUACCCAUUACACUGCCA -...(((.--.(((((..-((((--(.....((((((((((((((...........)))))))(.((((.....)))).).-.))))))).))))).....)))))...))). ( -42.60, z-score = -3.13, R) >droSec1.super_4 4391335 107 + 6179234 -ACUGGCC--AGUUAUUUUAGCC--GCUAAUGCGCGGAGUGCGGCCAACAAAAGCCGCCGCACGCGCUGACCUCCAGCUCA-GUCCGUUUCCGGCUUACCCAGUACACUGCCA -...(((.--(((((((..((((--((....))((((((((((((...........)))))))(.((((.....)))).).-.)))))....)))).....)))).)))))). ( -39.90, z-score = -2.31, R) >droYak2.chr3R 7898733 106 - 28832112 -ACUGGCC--AGUAAUUU-AGCC--GCUAAUGCGCGGAGUGCGGCCAACAAAAGCCGCCGCACGCGCUGACCUCCAGCUCA-GUCCGUGUCCGGCUUACCCAGUACACUGCCC -(((((..--.(((....-((((--(.....((((((((((((((...........)))))))(.((((.....)))).).-.))))))).)))))))))))))......... ( -41.00, z-score = -2.40, R) >droEre2.scaffold_4820 8006350 106 - 10470090 -ACUGGCC--AGUAAUUU-AGCC--GCUAAUGCGCGGAGUGCGGCCAACAAAAGCCGCCGCACGCGCUGACCUCCAGCUCA-GUCCGCGUCCGGCUUGGCCAGUACACUGCUA -(((((((--(.......-((((--(.....((((((((((((((...........)))))))(.((((.....)))).).-.))))))).)))))))))))))......... ( -50.41, z-score = -3.65, R) >droPer1.super_7 2574103 98 + 4445127 AACUGGUC--CGUAAUUU-AGCCACGCUAAUGCGCGGAGUGCGGUCAACAAAAGCCACAGCACGCGCUGACCUCCAGCUCCUGUCCACCUCCACGUCCGCG------------ ........--.(((..((-(((...))))))))((((((((.(((..(((..(((..((((....)))).......)))..)))..)))..))).))))).------------ ( -27.30, z-score = -0.58, R) >droVir3.scaffold_12855 1097656 102 + 10161210 CACAGACCUUAGUAAUUU-AGCCACGCUAAUGCGCGGACUGUGGUCAACAAAAGCCGCCGCACGCGCCGCGUUACUUCCCAC-----CGCCCAAC-CGCACCACCCUGC---- ..(((.....((((((((-(((...))))).(((((..(.(((((........))))).)..)))))...))))))......-----.((.....-.))......))).---- ( -25.20, z-score = -0.49, R) >droMoj3.scaffold_6540 26852929 98 + 34148556 CACAGACCUUAGUUAUUU-AGCCACGCUAAUGCGCGGACUGCGGUCAACAAAAGCAGCCGCACGCGCUCA-----CCACAAC-----CACCCAUAGUG-GCCACCCAACC--- ..................-.(((((((....))((((.((((...........)))))))).........-----.......-----........)))-)).........--- ( -25.00, z-score = -1.60, R) >droGri2.scaffold_14624 3679676 108 + 4233967 CACUGAUUUUAGUAAUUU-AGCCACGCUAAUGCGCGGACUGUGGUCAACAAAAGCCGUCGCACGCGCUGAAAUGCCCACUACAAAACCACCUAACACACACCACCCAAC---- ...((.((((.(((....-((((.(((....))).)).))((((.((.....(((((.....)).)))....)).))))))))))).))....................---- ( -20.40, z-score = -0.39, R) >consensus _ACUGGCC__AGUAAUUU_AGCC__GCUAAUGCGCGGAGUGCGGUCAACAAAAGCCGCCGCACGCGCUGACCUCCAGCUCA_GUCCGCGUCCGGCUUACCCAGUACACUG___ ...................(((...)))...(((((....(((((........)))))....))))).............................................. (-16.60 = -16.33 + -0.27)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:55:18 2011