
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 25,467,186 – 25,467,305 |
| Length | 119 |
| Max. P | 0.627073 |

| Location | 25,467,186 – 25,467,305 |
|---|---|
| Length | 119 |
| Sequences | 15 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 75.22 |
| Shannon entropy | 0.60454 |
| G+C content | 0.55266 |
| Mean single sequence MFE | -40.31 |
| Consensus MFE | -20.80 |
| Energy contribution | -21.49 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.82 |
| Mean z-score | -0.80 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.627073 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
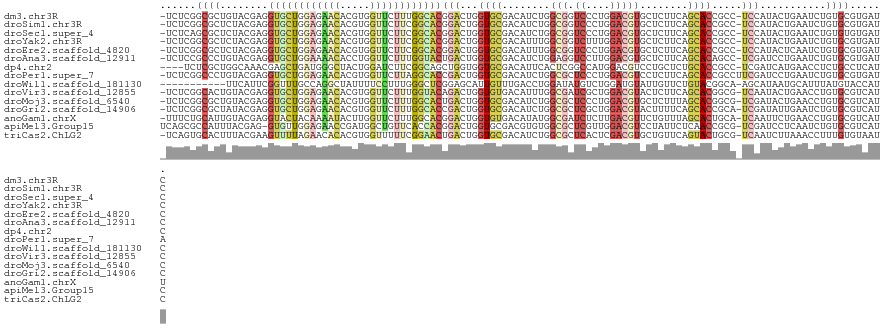
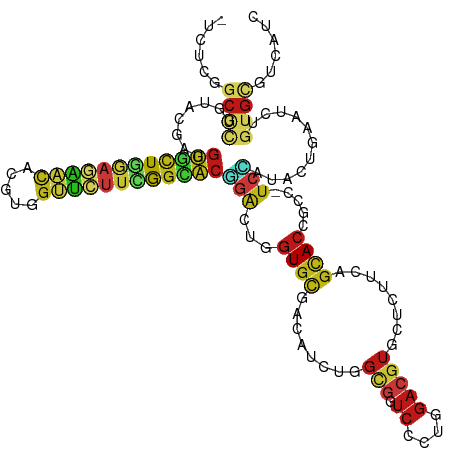
>dm3.chr3R 25467186 119 - 27905053 -UCUCGGCGCUGUACGAGGUGCUGGAGAACACGUGGUUCUUUGGCACGGACUGGUGCGACAUCUGGCGGUCCCUGGACGUGCUCUUCAGCACCGCC-UCCAUACUGAAUCUGUGCGUGAUC -..((((((((((((.((((((..((((((.....))))))..))))...)).)))))......(((((((....)))(((((....)))))))))-..............)))).))).. ( -45.80, z-score = -1.01, R) >droSim1.chr3R 25140163 119 - 27517382 -UCUCGGCGCUCUACGAGGUGCUGGAGAACACGUGGUUCUUCGGCACGGACUGGUGCGACAUCUGGCGGUCCCUGGACGUGCUCUUCAGCACCGCC-UCCAUACUGAAUCUGUGCGUGAUC -..(((((((......((((((((((((((.....))))))))))))(((..((((...)))).(((((((....)))(((((....)))))))))-)))...))......)))).))).. ( -45.30, z-score = -0.85, R) >droSec1.super_4 4323891 119 - 6179234 -UCUCAGCGCUCUACGAGGUGCUGGAGAACACGUGGUUCUUCGGCACGGACUGGUGCGACAUCUGGCGGUCCCUGGACGUGCUCUUCAGCACCGCC-UCCAUACUGAAUCUGUGUGUGAUC -.....((((.(((....((((((((((((.....))))))))))))....))).))).)....(((((((....)))(((((....)))))))))-(((((((.......))))).)).. ( -46.30, z-score = -1.39, R) >droYak2.chr3R 7825753 119 + 28832112 -UCUCGGCGCUCUACGAGGUGCUGGAGAACACGUGGUUCUUCGGCACGGACUGGUGCGACAUUUGGCGGUCUUUGGACGUGCUCUUCAGCACCGCC-UCCAUACUGAAUCUGUGCGUGAUC -..(((((((......((((((((((((((.....))))))))))))(((.((((((.......(((((((....))).)))).....))))))..-)))...))......)))).))).. ( -44.10, z-score = -1.11, R) >droEre2.scaffold_4820 7937040 119 + 10470090 -UCUCGGCGCUCUACGAGGUGCUGGAGAACACGUGGUUCUUCGGCACGGACUGGUGCGACAUUUGGCGGUCCCUGGACGUGCUCUUCAGCACCGCC-UCCAUACUCAAUCUGUGCGUGAUC -..(((((((.....(((((((((((((((.....))))))))))))(((.((((((.......(((((((....))).)))).....))))))..-)))...))).....)))).))).. ( -48.50, z-score = -2.08, R) >droAna3.scaffold_12911 5162475 119 - 5364042 -UCUCCGCCCUGUACGAGGUGCUGGAAAACACCUGGUUCUUUGGUACUGACUGGUGCGACAUCUGGAGGUCCUUGGACGUGCUCUUCAGCACAGCC-UCGAUCCUGAAUCUGUGCGUGAUC -.((((((((.((....(((((..((.(((.....))).))..))))).)).)).))(....).))))(((....))).......(((((((((..-(((....)))..)))))).))).. ( -37.10, z-score = -0.18, R) >dp4.chr2 14799796 116 - 30794189 ----UCUCGCUGGCAAACGAGCUGAUGGGCUACUGGAUCUUCGGCAGCUGGUGGUGCGACAUUCACUCGGCCAUGGACGUCCUGCUCUGCACCGCC-UCGAUCAUGAACCUCUGCCUCAUC ----....(((.(....).))).(((((((....((.((.((((.....((((((((((.......))(((...((....)).)))..))))))))-))))....)).))...))).)))) ( -35.00, z-score = 0.46, R) >droPer1.super_7 2504729 120 - 4445127 -UCUCGGCCCUGUACGAGGUGCUGGAGAACACGUGGUUCUUAGGCACCGACUGGUGCGACAUCUGGCGCUCCCUGGACGUCCUCUUCAGCACCGCCUUCGAUCCUGAAUCUGUGCGUGAUA -..........(((((.(((((((((((...............(((((....)))))(((.((..(......)..)).))).)))))))))))...((((....))))..)))))...... ( -39.30, z-score = 0.22, R) >droWil1.scaffold_181130 8813153 109 - 16660200 -----------UUCAUUCGGUUUGCCAGGCUAUUUUCCUUUGGGCUCGGAGCAUUGUUUGACCUGGAUAUGUCUGGAUGUAUUGUUCUGUACGGCA-AGCAUAAUGCAUUUAUGUACCAUC -----------.......(((...((((((.....(((...(((.(((((......)))))))))))...))))))(((((((((.((........-)).)))))))))......)))... ( -27.00, z-score = -0.13, R) >droVir3.scaffold_12855 818016 119 - 10161210 -UCUCGGCACUGUACGAGGUGCUGGAGAACACGUGGUUCUUUGGUACAGACUGGUGUGACAUUUGGCGAUCGCUGGACGUACUCUUCAGCACGGCG-UCAAUACUGAACCUGUGCGUCAUC -.((((........))))((((..((((((.....))))))..)))).(((.((.(((.(.((..((....))..)).)))).))...((((((..-(((....)))..)))))))))... ( -42.60, z-score = -1.30, R) >droMoj3.scaffold_6540 21993588 119 + 34148556 -UCUCGGCGCUGUACGAGGUGCUGGAGAACACGUGGUUCUUUGGCACUGACUGGUGCGACAUCUGGCGCUCCCUGGACGUGCUCUUUAGCACGGCG-UCGAUACUGAACCUGUGCGUCAUC -....(((((.......(((((..((((((.....))))))..))))).((.(((.((..(((.(((((((....))((((((....)))))))))-)))))..)).))).)))))))... ( -49.20, z-score = -2.28, R) >droGri2.scaffold_14906 1924256 119 - 14172833 -UCUCGGCGCUAUACGAGGUGCUGGAGAACACGUGGUUCUUUGGCACCGACUGGUGCGACAUCUGGCGCUCGCUGGACGUACUUUUCAGCACCGCA-UCGAUAUUGAAUCUGUGCGUCAUC -....((.(((......(((((..((((((.....))))))..)))))((..((((((...((..((....))..))))))))..))))).))(((-(.(((.....))).))))...... ( -50.10, z-score = -3.52, R) >anoGam1.chrX 10326760 119 + 22145176 -UUUCUGCAUUGUACGAGGUACUACAAAAUACUUGGUUCUUUGGCACGGACUGGUGUGACAUAUGGCGAUCUCUUGACGUUCUGUUUAGCACUGCA-UCAAUUCUGAACCUGUGCGUCAUU -......((..(.((.(((((........))))).)).)..))((((((...(((((...(((..(((.((....)))))..)))...)))))...-(((....)))..))))))...... ( -26.80, z-score = 0.36, R) >apiMel3.Group15 3247728 119 - 7856270 UCAGCGCCAUUUACGAG-GUGUUGGAGAACCGAUGGCUGUUCACCACGGACUGGUGCGACGUGUGGCGCUCGUUGGACGUCCUAUUCUCAACCGCG-UCGAUCCUCAAUCUGUGCGUCAUC ...((((((......((-.((((((....)))))).))((((.....))))))))))((((..((((((..((((((........)).)))).)))-))(((.....))).)..))))... ( -37.80, z-score = 0.64, R) >triCas2.ChLG2 4599585 119 - 12900155 -UCAGUGCACUUUACGAAGUUUUAGAACACACGUGGUUUUUCGGAACUGACUGGUGCGACAUCUGGCGCUCACUCGACGUGCUGUUCAGUACUGCG-UCAAUCUUAAACCUUUGUGUAAUC -.((((((((((....))))....(((((((((((((((....)))))((.((((((........)))).)).)).))))).))))).))))))..-........................ ( -29.70, z-score = 0.19, R) >consensus _UCUCGGCGCUGUACGAGGUGCUGGAGAACACGUGGUUCUUCGGCACGGACUGGUGCGACAUCUGGCGGUCCCUGGACGUGCUCUUCAGCACCGCC_UCCAUACUGAAUCUGUGCGUCAUC ......((((........((((((((((((.....))))))))))))(((...((((........(((.((....)))))........)))).....)))...........))))...... (-20.80 = -21.49 + 0.69)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:55:13 2011