
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 25,447,259 – 25,447,361 |
| Length | 102 |
| Max. P | 0.847725 |

| Location | 25,447,259 – 25,447,361 |
|---|---|
| Length | 102 |
| Sequences | 10 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 67.31 |
| Shannon entropy | 0.66667 |
| G+C content | 0.43023 |
| Mean single sequence MFE | -16.65 |
| Consensus MFE | -8.26 |
| Energy contribution | -8.94 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.40 |
| Mean z-score | -0.92 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.847725 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
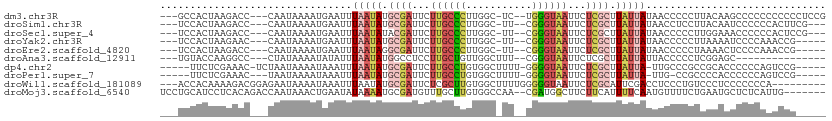
>dm3.chr3R 25447259 102 - 27905053 ---GCCACUAAGACC---CAAUAAAAUGAAUUUAAUAUGCGAUUCUUGCCCUUGGC-UC--UGGGUAAUUCUCGCUUAUUAUAACCCCCUUACAAGCCCCCCCCCCCUCCG ---((...((((...---((......))....(((((.((((...((((((.....-..--.))))))...)))).))))).......))))...)).............. ( -17.00, z-score = -2.28, R) >droSim1.chr3R 25119992 99 - 27517382 ---UCCACUAAGACC---CAAUAAAAUGAAUUUAAUAUGCGAUUCUUGCCCUUGGC-UU--CGGGUAAUUCUCGCUUAUUAUAACCUCCUUACAAUCCCCCCACUUCG--- ---............---..............(((((.((((...((((((.....-..--.))))))...)))).)))))...........................--- ( -15.40, z-score = -2.06, R) >droSec1.super_4 4303859 99 - 6179234 ---UCCACUAAGACC---CAAUAAAAUGAAUUUAAUAUACGAUUCUUGCCCUUGGC-UU--CGGGUAAUUCUCGCUUAUUAUAACCCCCUUGGAAACCCCCCACUCCG--- ---....(((((...---((......))....(((((..(((...((((((.....-..--.))))))...)))..))))).......)))))...............--- ( -15.20, z-score = -1.18, R) >droYak2.chr3R 7804051 97 + 28832112 ---UCCACUAAGAAC---CAAUAAAAUGAAUUUAAUAUGCGAUUCUUGCCCUUGGC-UU--CGGGUAAUUCUCGCUUAUUAUAACCCCCUUAAAAUCCCCAAACCG----- ---............---..............(((((.((((...((((((.....-..--.))))))...)))).))))).........................----- ( -15.40, z-score = -2.14, R) >droEre2.scaffold_4820 7916129 97 + 10470090 ---UCCACUAAGACC---CAAUAAAAUGAAUUUAAUAGGCGAUUCUUGCCCUUGGC-UU--CGGGUAAUUCUCGCUUAUUAUAACCCCCUAAAACUCCCCAAACCG----- ---............---..............((((((((((...((((((.....-..--.))))))...)))))))))).........................----- ( -19.20, z-score = -2.64, R) >droAna3.scaffold_12911 5141884 88 - 5364042 ---UGUACCAAGGCC---CUAUAAAAUAUAUUUAAUAUGGCCUCCUUGCUGUUGGCUUU--CGGGUAAUUCUCGCUUAUUAUUACCCCCUCGGAGC--------------- ---.(((...(((((---.(((.(((....))).))).)))))...))).....(((((--.(((((((...........)))))))....)))))--------------- ( -19.10, z-score = -0.26, R) >dp4.chr2 2319380 98 - 30794189 -----UUCUCGAAAC-UCUAAUAAAAUAAAUUUAAUAUGCGAUUCUUGCCUGUGGCUUUU-GGGGUAAUUCUCGCUUAUUA-UUGCCCGCCGCACCCCCCAGUCCG----- -----..........-......................(.((((......((((((....-.(((((((...........)-))))))))))))......))))).----- ( -16.50, z-score = -0.07, R) >droPer1.super_7 2484695 95 - 4445127 -----UUCUCGAAAC---UAAUAAAAUAAAUUUAAUAUGCGAUUCUUGCCUGUGGCUUUU-GGGGUAAUUCUCGCUUAUUA-UUG-CCGCCCCACCCCCCAGUCCG----- -----..........---................(((.((((...)))).)))((((...-(((((.......((......-..)-)......)))))..))))..----- ( -13.52, z-score = 0.69, R) >droWil1.scaffold_181089 5721738 99 - 12369635 ---ACCACAAAAGACGGAGAAUAAAAUAAAUUUAAUAUGCGAUUCUCGCUUGUGGCUUUUGGGGGUAAUUCUCGCAUUCGACCUCCCUGUCCCUCCCCCCCA--------- ---.(((((..((...((((((...(((.......)))...)))))).)))))))....((((((........(....)(((......))).....))))))--------- ( -21.10, z-score = -0.23, R) >droMoj3.scaffold_6540 21971418 102 + 34148556 UCCUGCAUCCUCACAGACCAAUAAACUGAAUAUAAAAUGCGAUGUUUGCUUGUGGCCAA--CGAUGGCUUCUUCAUUUUCAAUGUUUUCUGAAUGCUCUCAUUG------- ....((((..(((.((((....(((.((((........((((...))))..(.(((((.--...))))).))))).)))....))))..)))))))........------- ( -14.10, z-score = 0.93, R) >consensus ___UCCACUAAGACC___CAAUAAAAUGAAUUUAAUAUGCGAUUCUUGCCCUUGGC_UU__CGGGUAAUUCUCGCUUAUUAUAACCCCCUCACAACCCCCAAACCG_____ ................................(((((.((((...((((((...........))))))...)))).))))).............................. ( -8.26 = -8.94 + 0.68)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:55:11 2011