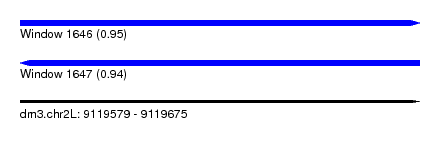
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 9,119,579 – 9,119,675 |
| Length | 96 |
| Max. P | 0.950917 |

| Location | 9,119,579 – 9,119,675 |
|---|---|
| Length | 96 |
| Sequences | 12 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 67.02 |
| Shannon entropy | 0.67986 |
| G+C content | 0.41804 |
| Mean single sequence MFE | -27.05 |
| Consensus MFE | -9.57 |
| Energy contribution | -9.49 |
| Covariance contribution | -0.07 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.950917 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
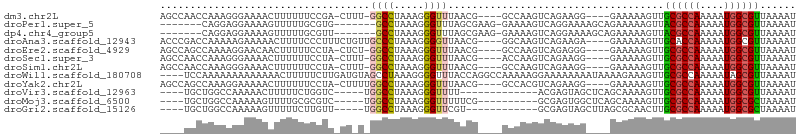
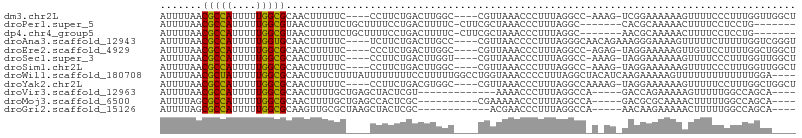
>dm3.chr2L 9119579 96 + 23011544 AGCCAACCAAAGGGAAAACUUUUUUCCGA-CUUU-GGCCUAAAGGGUUUAACG----GCCAAGUCAGAAGG----GAAAAAGUUGCGCCAAAAAUGGCGUUAAAAU .((((.......((..(((((((((((((-(((.-((((.............)----))))))))....))----)))))))))...)).....))))........ ( -31.72, z-score = -2.70, R) >droPer1.super_5 5829744 91 + 6813705 -------CAGGAGGAAAAGUUUUUGCGUG-------GCCUAAAGGGUUUAGCGAAG-GAAAAGUCAGGAAAAGCAGAAAAAGUUACGCCAAAAAUGGCGUUAAAAU -------............(((((((...-------.(((....(..((..(....-)..))..))))....))))))).....((((((....))))))...... ( -19.60, z-score = -1.76, R) >dp4.chr4_group5 1423404 91 + 2436548 -------CAGGAGGAAAAGUUUUUGCGUU-------GCCUAAAGGGUUUAGCGAAG-GAAAAGUCAGGAAAAGCAGAAAAAGUUACGCCAAAAAUGGCGUUAAAAU -------............(((((((...-------.(((....(..((..(....-)..))..))))....))))))).....((((((....))))))...... ( -19.60, z-score = -1.58, R) >droAna3.scaffold_12943 468124 98 + 5039921 ACCCGACCAAAAAGAAAAACUUUUCCCUUUCUGUUGCCCUAAAGGGGUUAACG----GGCAAGUCAGAAGA----GAAAAAGUUGCACCAAAAAUGGCGUUAAAAU ...((.(((........(((((((.(.((((((((((((.............)----)))))..)))))).----).)))))))..........)))))....... ( -21.99, z-score = -0.65, R) >droEre2.scaffold_4929 9724101 96 + 26641161 AGCCAGCCAAAAGGAACAACUUUUUCCUA-CUCU-GGCCUAAAGGGUUUAACG----GCCAAGUCAGAGGG----GAAAAAGUUGCGCCAAAAAUGGCGUUAAAAU .((((.......((..((((((((((((.-((((-(((......(((......----)))..)))))))))----))))))))))..)).....))))........ ( -37.70, z-score = -4.28, R) >droSec1.super_3 4585714 96 + 7220098 AGCCAACCAAAGGGAAAACUUUUUUCCUA-CUUU-GGCCUAAAGGGUUUAACG----ACCAAGUCAGAAGG----GAAAAAGUUGCGCCAAAAAUGGCGUUAAAAU .((((.......((..((((((((((((.-.(((-(((......((((....)----)))..)))))))))----)))))))))...)).....))))........ ( -29.20, z-score = -2.51, R) >droSim1.chr2L 8899685 96 + 22036055 AGCCAACCAAAGGGAAAACUUUUUUCCUA-CUUU-GGCCUAAAGGGUUUAACG----GCCAAGUCAGAAGG----GAAAAAGUUGCGCCAAAAAUGGCGUUAAAAU .((((.......((..((((((((((((.-((((-((((.............)----)))))...)).)))----)))))))))...)).....))))........ ( -31.32, z-score = -2.62, R) >droWil1.scaffold_180708 49506 102 + 12563649 ----UCCAAAAAAAAAAAAACUUUUUCUUGAUGUAGCCUAAAGGGGUUUACCAGGCCAAAAAGGAAAAAAAAUAAAAGAAAGUUGCGCCAAAAAUAGCGUUAAAAU ----.................((((((((..((..((((...((......))))))))..))))))))................((((........))))...... ( -15.40, z-score = -0.19, R) >droYak2.chr2L 11786006 97 + 22324452 AGCCAGCCAAAGGAAAAACUUUUUUCCUA-CUUUUGGCCUAAAGGGUUUAACG----GCCACGUCAGAAGG----GAAAAAGUUGCGCCAAAAAUGGCGUUAAAAU .((((......((...((((((((((((.-((..(((((.............)----))))....)).)))----)))))))))...)).....))))........ ( -31.12, z-score = -2.42, R) >droVir3.scaffold_12963 467998 84 + 20206255 ----UGCUGGCCAAAAACUUUUUCUGGUC-----UGGCCUAAAGGGUUUU-------------ACGAGUAGCUCAGCAAAAGUUGCGCCAAAAAUGGCGUUAAAAU ----(((((((((...(((......))).-----)))))....(((((..-------------......)))))))))......((((((....))))))...... ( -24.70, z-score = -1.49, R) >droMoj3.scaffold_6500 11968325 87 + 32352404 ----UGCUGGCCAAAAAGUUUUGCGCGUC-----UGGCCUAAAGGGUUUUUCG----------GCGAGUGGCUCAGCAAAAGUUGCGCCAAAAAUGGCGCUAAAAU ----(((((((((......(((((.((..-----.((((.....))))...))----------))))))))).)))))......((((((....))))))...... ( -33.20, z-score = -2.09, R) >droGri2.scaffold_15126 5250620 85 - 8399593 ----UGCUGGCCAAAAAGUUUUUCUUGUU-----UGGCCUAAAGGGUUCGU------------GCGAGUAGCUUAGCGCAACUUGCGCCAAAAAUGGCGCUAAAAU ----....((((((((((.....))).))-----)))))....(((((.((------------((..........)))))))))((((((....))))))...... ( -29.10, z-score = -1.70, R) >consensus ____AGCCAAAAGGAAAACUUUUUUCCUA_CU_U_GGCCUAAAGGGUUUAACG____GCCAAGUCAGAAGG____GAAAAAGUUGCGCCAAAAAUGGCGUUAAAAU ...................................((((.....))))....................................((((((....))))))...... ( -9.57 = -9.49 + -0.07)
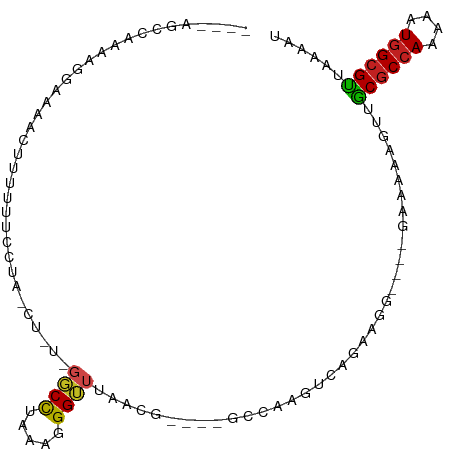
| Location | 9,119,579 – 9,119,675 |
|---|---|
| Length | 96 |
| Sequences | 12 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 67.02 |
| Shannon entropy | 0.67986 |
| G+C content | 0.41804 |
| Mean single sequence MFE | -24.88 |
| Consensus MFE | -7.39 |
| Energy contribution | -7.23 |
| Covariance contribution | -0.15 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.940239 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 9119579 96 - 23011544 AUUUUAACGCCAUUUUUGGCGCAACUUUUUC----CCUUCUGACUUGGC----CGUUAAACCCUUUAGGCC-AAAG-UCGGAAAAAAGUUUUCCCUUUGGUUGGCU .......(((((....)))))(((((.....----..((((((((((((----(.............))))-.)))-)))))).((((......)))))))))... ( -30.12, z-score = -3.03, R) >droPer1.super_5 5829744 91 - 6813705 AUUUUAACGCCAUUUUUGGCGUAACUUUUUCUGCUUUUCCUGACUUUUC-CUUCGCUAAACCCUUUAGGC-------CACGCAAAAACUUUUCCUCCUG------- ......((((((....)))))).........(((....(((((......-..............))))).-------...)))................------- ( -13.65, z-score = -2.41, R) >dp4.chr4_group5 1423404 91 - 2436548 AUUUUAACGCCAUUUUUGGCGUAACUUUUUCUGCUUUUCCUGACUUUUC-CUUCGCUAAACCCUUUAGGC-------AACGCAAAAACUUUUCCUCCUG------- ......((((((....)))))).........(((.((.(((((......-..............))))).-------)).)))................------- ( -14.65, z-score = -2.63, R) >droAna3.scaffold_12943 468124 98 - 5039921 AUUUUAACGCCAUUUUUGGUGCAACUUUUUC----UCUUCUGACUUGCC----CGUUAACCCCUUUAGGGCAACAGAAAGGGAAAAGUUUUUCUUUUUGGUCGGGU .......(((((.....((...(((((((((----..(((((..(((((----(.............)))))))))))..)))))))))...))...))).))... ( -30.12, z-score = -2.56, R) >droEre2.scaffold_4929 9724101 96 - 26641161 AUUUUAACGCCAUUUUUGGCGCAACUUUUUC----CCCUCUGACUUGGC----CGUUAAACCCUUUAGGCC-AGAG-UAGGAAAAAGUUGUUCCUUUUGGCUGGCU ........((((.....((.(((((((((((----(.((((.....(((----(.............))))-))))-..)))))))))))).))...))))..... ( -34.02, z-score = -3.55, R) >droSec1.super_3 4585714 96 - 7220098 AUUUUAACGCCAUUUUUGGCGCAACUUUUUC----CCUUCUGACUUGGU----CGUUAAACCCUUUAGGCC-AAAG-UAGGAAAAAAGUUUUCCCUUUGGUUGGCU ........((((.....((...((((((((.----(((.((...(((((----(.............))))-))))-.))).))))))))...))......)))). ( -24.12, z-score = -1.16, R) >droSim1.chr2L 8899685 96 - 22036055 AUUUUAACGCCAUUUUUGGCGCAACUUUUUC----CCUUCUGACUUGGC----CGUUAAACCCUUUAGGCC-AAAG-UAGGAAAAAAGUUUUCCCUUUGGUUGGCU ........((((.....((...((((((((.----(((.((...(((((----(.............))))-))))-.))).))))))))...))......)))). ( -26.82, z-score = -1.82, R) >droWil1.scaffold_180708 49506 102 - 12563649 AUUUUAACGCUAUUUUUGGCGCAACUUUCUUUUAUUUUUUUUCCUUUUUGGCCUGGUAAACCCCUUUAGGCUACAUCAAGAAAAAGUUUUUUUUUUUUUGGA---- .......(((((....)))))...........................((((((((.........))))))))..(((((((((((....))))))))))).---- ( -20.30, z-score = -1.89, R) >droYak2.chr2L 11786006 97 - 22324452 AUUUUAACGCCAUUUUUGGCGCAACUUUUUC----CCUUCUGACGUGGC----CGUUAAACCCUUUAGGCCAAAAG-UAGGAAAAAAGUUUUUCCUUUGGCUGGCU ........((((.((..((...((((((((.----(((.((....((((----(.............)))))..))-.))).))))))))...))...)).)))). ( -27.52, z-score = -1.68, R) >droVir3.scaffold_12963 467998 84 - 20206255 AUUUUAACGCCAUUUUUGGCGCAACUUUUGCUGAGCUACUCGU-------------AAAACCCUUUAGGCCA-----GACCAGAAAAAGUUUUUGGCCAGCA---- ........((((....))))((...((((((.(((...)))))-------------)))).......(((((-----((............))))))).)).---- ( -22.10, z-score = -1.63, R) >droMoj3.scaffold_6500 11968325 87 - 32352404 AUUUUAGCGCCAUUUUUGGCGCAACUUUUGCUGAGCCACUCGC----------CGAAAAACCCUUUAGGCCA-----GACGCGCAAAACUUUUUGGCCAGCA---- ......((((((....))))))......(((((.((((((.((----------(.(((.....))).))).)-----)....(.....)....)))))))))---- ( -27.20, z-score = -2.01, R) >droGri2.scaffold_15126 5250620 85 + 8399593 AUUUUAGCGCCAUUUUUGGCGCAAGUUGCGCUAAGCUACUCGC------------ACGAACCCUUUAGGCCA-----AACAAGAAAAACUUUUUGGCCAGCA---- ......((((((....)))))).......(((..((.....))------------............(((((-----((.(((.....))))))))))))).---- ( -27.90, z-score = -2.72, R) >consensus AUUUUAACGCCAUUUUUGGCGCAACUUUUUC____CCUUCUGACUUGGC____CGUUAAACCCUUUAGGCC_A_AG_UAGGAAAAAAGUUUUCCUUUUGGCU____ .......(((((....)))))..................................................................................... ( -7.39 = -7.23 + -0.15)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:27:35 2011