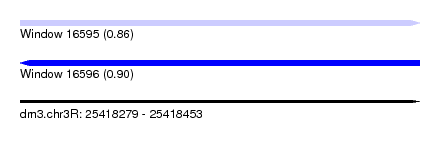
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 25,418,279 – 25,418,453 |
| Length | 174 |
| Max. P | 0.904916 |

| Location | 25,418,279 – 25,418,453 |
|---|---|
| Length | 174 |
| Sequences | 4 |
| Columns | 192 |
| Reading direction | forward |
| Mean pairwise identity | 85.05 |
| Shannon entropy | 0.23643 |
| G+C content | 0.44603 |
| Mean single sequence MFE | -54.00 |
| Consensus MFE | -39.66 |
| Energy contribution | -39.73 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.856076 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
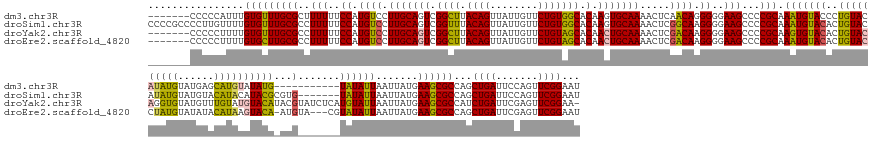
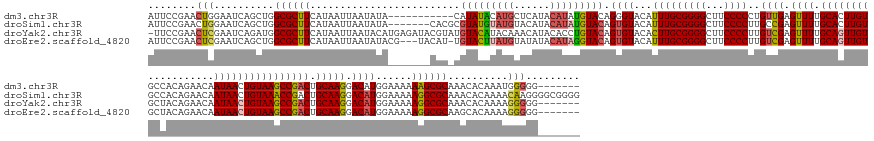
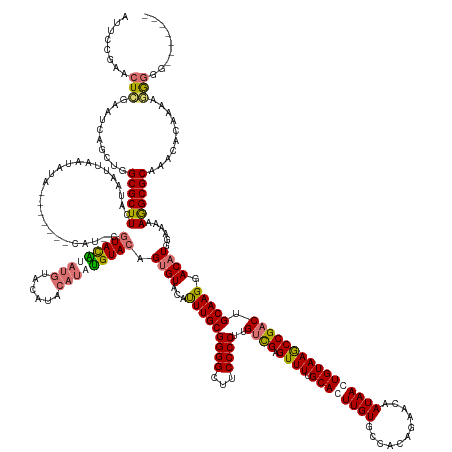
>dm3.chr3R 25418279 174 + 27905053 -------CCCCCAUUUGUGUUUGCGCUUUUUUCCAUGUCCUUGCAGUCGGCUUACAGUUAUUGUUCUGUGGCACAAGUGCAAAACUCAACAGGGGGAAGCCCCGCAAAUGUACCCUGUACAUAUGUAUGAGCAUGUAUAUG-----------UAUAUUAAUUAUGAAGCGCCAGCUGAUUCCAGUUCGGAAU -------(((((..(((..(((((((((....((((......(((((.(((.....))))))))...))))...)))))))))...)))..))))).(((..(((..(((.....(((((((((((((....)))))))))-----------)))).....)))...)))...))).(((((.....))))) ( -48.60, z-score = -0.93, R) >droSim1.chr3R 25090900 185 + 27517382 CCCCGCCCCUUGUUUUGUGUUUGCGCCUUUUUCCAUGUCCUUGCAGUCGGUUUACAGUUAUUGUUCUGUGGCACAAGUGCAAAACUCGGCAAGGGGAAGCCCCGCAAAUGUACACUGUACAUAUGUAUGUACAUACAUACGCGUG-------UAUAUUAAUUAUGAAGCGCCAGCUGAUUCCAGUUCGGAAU ((((...((..((((((..((((.(((.....((.(((....)))...))...((((........))))))).))))..))))))..))...)))).(((..(((..(((.....((((((..(((((((....)))))))..))-------)))).....)))...)))...))).(((((.....))))) ( -57.20, z-score = -2.10, R) >droYak2.chr3R 7774340 184 - 28832112 -------CCCCCUUUUGUGUUUGCGCCUUUUUCCAUGUCCUUGCAGUCGGCUUACAGUUAUUGUUCUGUAGCACAACUGCAAAACUCGACAAGGGGAAGCCCCGCAAGUGUACACUGUACAGGUGUAUGUUUGUAUGUACAUACGUAUCUCAUGUAUUAAUUAUGAAGCGCCAUCUGAUUCGAGUUCGGAA- -------.........(((....)))....((((......(((((((.((((.((((........))))))).).)))))))(((((((...((((...))))(((((..((((((.....))))))..))))).........(((...(((((.......))))).))).........))))))).))))- ( -52.80, z-score = -1.67, R) >droEre2.scaffold_4820 7887932 181 - 10470090 -------CCCCCUUUUGUGCUUGCGCCUUUUUCCAUGUCCUUGCAGUCGGCUUACAGUUAUUGUUCUGUAGCACAACUGCAAAACUCGACAAGGGGAAGCCCCGCAAAUGUACACUGUACCUAUGUAUAUACAUAAGUACA-AUGUA---CGUAUAUUAAUUAUGAAGCGCCAGCUGAUUCGAGUUCGGAAU -------.........(((((((((..(((..((.((((.(((((((.((((.((((........))))))).).))))))).....)))).))..)))...)))..(((((((.(((((.(((((....))))).)))))-.))))---)))............))))))...((((.......))))... ( -57.40, z-score = -3.89, R) >consensus _______CCCCCUUUUGUGUUUGCGCCUUUUUCCAUGUCCUUGCAGUCGGCUUACAGUUAUUGUUCUGUAGCACAACUGCAAAACUCGACAAGGGGAAGCCCCGCAAAUGUACACUGUACAUAUGUAUGUACAUAUAUACA_AUG_______UAUAUUAAUUAUGAAGCGCCAGCUGAUUCCAGUUCGGAAU ................(((((((((..(((..((.((((.(((((((.((((.((((........))))))).).))))))).....)))).))..)))...)))..........((((((((((......))))))))))........................))))))...((((.......))))... (-39.66 = -39.73 + 0.06)
| Location | 25,418,279 – 25,418,453 |
|---|---|
| Length | 174 |
| Sequences | 4 |
| Columns | 192 |
| Reading direction | reverse |
| Mean pairwise identity | 85.05 |
| Shannon entropy | 0.23643 |
| G+C content | 0.44603 |
| Mean single sequence MFE | -54.84 |
| Consensus MFE | -41.06 |
| Energy contribution | -42.12 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.904916 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
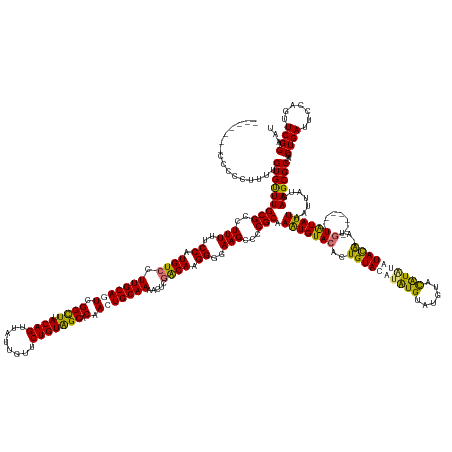
>dm3.chr3R 25418279 174 - 27905053 AUUCCGAACUGGAAUCAGCUGGCGCUUCAUAAUUAAUAUA-----------CAUAUACAUGCUCAUACAUAUGUACAGGGUACAUUUGCGGGGCUUCCCCCUGUUGAGUUUUGCACUUGUGCCACAGAACAAUAACUGUAAGCCGACUGCAAGGACAUGGAAAAAAGCGCAAACACAAAUGGGGG------- .(((((..(((....)))...((((((.(((.....))).-----------......((((((((..((.((((((...)))))).)).((((....))))...))))(((((((.(((.((.((((........))))..))))).))))))).)))).....)))))).........))))).------- ( -41.30, z-score = 0.69, R) >droSim1.chr3R 25090900 185 - 27517382 AUUCCGAACUGGAAUCAGCUGGCGCUUCAUAAUUAAUAUA-------CACGCGUAUGUAUGUACAUACAUAUGUACAGUGUACAUUUGCGGGGCUUCCCCUUGCCGAGUUUUGCACUUGUGCCACAGAACAAUAACUGUAAACCGACUGCAAGGACAUGGAAAAAGGCGCAAACACAAAACAAGGGGCGGGG ..((((..((.........((((((.........(((.((-------(((..((((((((((....)))))))))).))))).))).))((((...))))..)))).((((((...(((((((((((........))))...((........))...........)))))))...))))))..))..)))). ( -55.60, z-score = -1.65, R) >droYak2.chr3R 7774340 184 + 28832112 -UUCCGAACUCGAAUCAGAUGGCGCUUCAUAAUUAAUACAUGAGAUACGUAUGUACAUACAAACAUACACCUGUACAGUGUACACUUGCGGGGCUUCCCCUUGUCGAGUUUUGCAGUUGUGCUACAGAACAAUAACUGUAAGCCGACUGCAAGGACAUGGAAAAAGGCGCAAACACAAAAGGGGG------- -.......(((..........((((((.....................((((((........)))))).((((((.(((....)))))))))..((((...((((....((((((((((.(((((((........)))).))))))))))))))))).))))..))))))..........)))..------- ( -53.45, z-score = -2.12, R) >droEre2.scaffold_4820 7887932 181 + 10470090 AUUCCGAACUCGAAUCAGCUGGCGCUUCAUAAUUAAUAUACG---UACAU-UGUACUUAUGUAUAUACAUAGGUACAGUGUACAUUUGCGGGGCUUCCCCUUGUCGAGUUUUGCAGUUGUGCUACAGAACAAUAACUGUAAGCCGACUGCAAGGACAUGGAAAAAGGCGCAAGCACAAAAGGGGG------- .((((((.......)).(((.((((((..............(---(((((-(((((((((((....))))))))))))))))).(((.(((((...)))).((((....((((((((((.(((((((........)))).))))))))))))))))).).))).)))))).)))......)))).------- ( -69.00, z-score = -5.77, R) >consensus AUUCCGAACUCGAAUCAGCUGGCGCUUCAUAAUUAAUAUA_______CAU_CGUACAUAUGUACAUACAUAUGUACAGUGUACAUUUGCGGGGCUUCCCCUUGUCGAGUUUUGCACUUGUGCCACAGAACAAUAACUGUAAGCCGACUGCAAGGACAUGGAAAAAGGCGCAAACACAAAAGGGGG_______ ........(((..........((((((.........................(((((((((......))))))))).((((...(((((((((...))))..((((.((((.((((((((...........)))))))))))))))).))))).))))......))))))..........)))......... (-41.06 = -42.12 + 1.06)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:55:10 2011