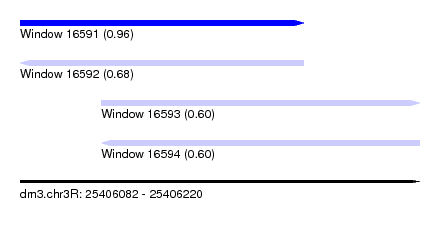
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 25,406,082 – 25,406,220 |
| Length | 138 |
| Max. P | 0.955004 |

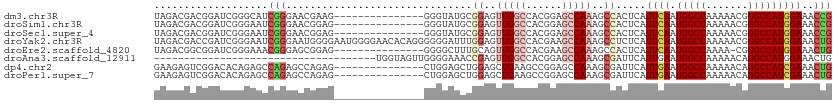
| Location | 25,406,082 – 25,406,180 |
|---|---|
| Length | 98 |
| Sequences | 8 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 70.52 |
| Shannon entropy | 0.57149 |
| G+C content | 0.56205 |
| Mean single sequence MFE | -32.84 |
| Consensus MFE | -15.57 |
| Energy contribution | -16.39 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.955004 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
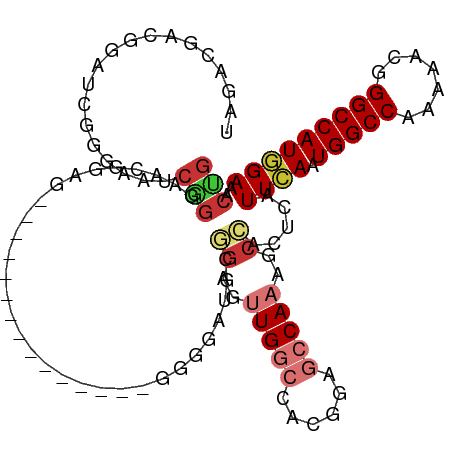
>dm3.chr3R 25406082 98 + 27905053 UAGACGACGGAUCGGGCAUCGGGAACGAAG---------------GGGUAUGCGGAGUUGGCCACGGAGCCAAAGCCACUCAUUCAAUGGCCAAAAACGGGCCAUGGAAACCG .......(((...(((..(((....)))..---------------........((..(((((......)))))..)).))).(((.((((((.......)))))).))).))) ( -31.40, z-score = -0.95, R) >droSim1.chr3R 25078399 98 + 27517382 UAGACGACGGAUCGGGAAUCGGGAACGGAG---------------GGGUAUGCGGAGUUGGCCACGGAGCCAAAGCCACUCAUUCAAUGGCCAAAAACGGGCCAUGGAAACCG .......(((...(((..(((....)))..---------------........((..(((((......)))))..)).))).(((.((((((.......)))))).))).))) ( -31.20, z-score = -0.92, R) >droSec1.super_4 4263723 98 + 6179234 UAGACGACGGAUCGGGAAUCGGGAACGGAG---------------GGGUAUGCGGAGUUGGCCACGGAGCCAAAGCCACUCAUUCAAUGGCCAAAAACGGGCCAUGGAAACCG .......(((...(((..(((....)))..---------------........((..(((((......)))))..)).))).(((.((((((.......)))))).))).))) ( -31.20, z-score = -0.92, R) >droYak2.chr3R 7761038 113 - 28832112 UAGACGACCGAUCGGGAAUCGGGAAUGGGGAAUGGGGAACACAGGGGGGAUUUGGAGUUGGCCACGAAGCCAAAGCCUCUCAUUCAAUGGCCAAAAACGGGCCAUGGAAACUG .......(((((.....)))))((((......(((....).))((((((.(((((..(((....)))..))))).)))))))))).((((((.......))))))(....).. ( -36.80, z-score = -1.50, R) >droEre2.scaffold_4820 7873684 97 - 10470090 UAGACGGCGGAUCGGGAAACGGGAGCGGAG---------------GGGGCUUUGCAGUUGGCCACGAAGCCAAAGCCACUCAUUCAAUGGCCAAAA-CGGGCCAUGGAAACUG ....(.((...((.(....).)).)).).(---------------(((((((((...(((....)))...))))))).))).(((.((((((....-..)))))).))).... ( -34.40, z-score = -1.69, R) >droAna3.scaffold_12911 5101465 76 + 5364042 -------------------------------------UGGUAGUUGGGGAAACCGAGUUGGCCACGGAGCCAAAGCGAUUCAUUGAAUGGCCAAAAACAGGCCAUGGAAACUG -------------------------------------...(((((.((....))(..(((((......)))))..)..........((((((.......))))))...))))) ( -23.30, z-score = -1.47, R) >dp4.chr2 2276653 98 + 30794189 GAAGAGUCGGACACAGAGCCAGAGCCAGAG---------------CUGGAGCUGGAGCUGAAGCCGGAGCCAAAGCGAUUCAUUGAAUGGCCAAAAACAGGCCAUCGAAACUG ...(((((((...(((..((((..(((...---------------.)))..))))..)))...))...((....))))))).((((.(((((.......)))))))))..... ( -37.20, z-score = -3.67, R) >droPer1.super_7 2441529 98 + 4445127 GAAGAGUCGGACACAGAGCCAGAGCCAGAG---------------CUGGAGCUGGAGCUGAAGCCGGAGCCAAAGCGAUUCAUUGAAUGGCCAAAAACAGGCCAUCGAAACUG ...(((((((...(((..((((..(((...---------------.)))..))))..)))...))...((....))))))).((((.(((((.......)))))))))..... ( -37.20, z-score = -3.67, R) >consensus UAGACGACGGAUCGGGAAUCGGGAACGGAG_______________GGGGAUGCGGAGUUGGCCACGGAGCCAAAGCCACUCAUUCAAUGGCCAAAAACGGGCCAUGGAAACUG ...................(((...............................((..(((((......)))))..)).....((((.(((((.......)))))))))..))) (-15.57 = -16.39 + 0.81)
| Location | 25,406,082 – 25,406,180 |
|---|---|
| Length | 98 |
| Sequences | 8 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 70.52 |
| Shannon entropy | 0.57149 |
| G+C content | 0.56205 |
| Mean single sequence MFE | -27.88 |
| Consensus MFE | -14.09 |
| Energy contribution | -14.12 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.682735 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 25406082 98 - 27905053 CGGUUUCCAUGGCCCGUUUUUGGCCAUUGAAUGAGUGGCUUUGGCUCCGUGGCCAACUCCGCAUACCC---------------CUUCGUUCCCGAUGCCCGAUCCGUCGUCUA .((((((.((((((.......)))))).)))...((((..((((((....))))))..))))..))).---------------.........(((((.......))))).... ( -29.40, z-score = -1.01, R) >droSim1.chr3R 25078399 98 - 27517382 CGGUUUCCAUGGCCCGUUUUUGGCCAUUGAAUGAGUGGCUUUGGCUCCGUGGCCAACUCCGCAUACCC---------------CUCCGUUCCCGAUUCCCGAUCCGUCGUCUA (((.(((.((((((.......)))))).)))...((((..((((((....))))))..))))......---------------........))).....(((....))).... ( -28.10, z-score = -0.94, R) >droSec1.super_4 4263723 98 - 6179234 CGGUUUCCAUGGCCCGUUUUUGGCCAUUGAAUGAGUGGCUUUGGCUCCGUGGCCAACUCCGCAUACCC---------------CUCCGUUCCCGAUUCCCGAUCCGUCGUCUA (((.(((.((((((.......)))))).)))...((((..((((((....))))))..))))......---------------........))).....(((....))).... ( -28.10, z-score = -0.94, R) >droYak2.chr3R 7761038 113 + 28832112 CAGUUUCCAUGGCCCGUUUUUGGCCAUUGAAUGAGAGGCUUUGGCUUCGUGGCCAACUCCAAAUCCCCCCUGUGUUCCCCAUUCCCCAUUCCCGAUUCCCGAUCGGUCGUCUA (((.(((.((((((.......)))))).))).....((..((((((....))))))..)).........)))...................((((((...))))))....... ( -27.50, z-score = -0.55, R) >droEre2.scaffold_4820 7873684 97 + 10470090 CAGUUUCCAUGGCCCG-UUUUGGCCAUUGAAUGAGUGGCUUUGGCUUCGUGGCCAACUGCAAAGCCCC---------------CUCCGCUCCCGUUUCCCGAUCCGCCGUCUA ....(((.((((((..-....)))))).))).((((((....(((((.(..(....)..).)))))..---------------..))))))...................... ( -29.80, z-score = -1.59, R) >droAna3.scaffold_12911 5101465 76 - 5364042 CAGUUUCCAUGGCCUGUUUUUGGCCAUUCAAUGAAUCGCUUUGGCUCCGUGGCCAACUCGGUUUCCCCAACUACCA------------------------------------- .((((...((((((.......)))))).....((((((..((((((....))))))..))))))....))))....------------------------------------- ( -22.10, z-score = -2.03, R) >dp4.chr2 2276653 98 - 30794189 CAGUUUCGAUGGCCUGUUUUUGGCCAUUCAAUGAAUCGCUUUGGCUCCGGCUUCAGCUCCAGCUCCAG---------------CUCUGGCUCUGGCUCUGUGUCCGACUCUUC .((..(((((((((.......))))))..........(((..((((..((((..(((....)))..))---------------))..))))..)))........)))..)).. ( -29.00, z-score = -0.72, R) >droPer1.super_7 2441529 98 - 4445127 CAGUUUCGAUGGCCUGUUUUUGGCCAUUCAAUGAAUCGCUUUGGCUCCGGCUUCAGCUCCAGCUCCAG---------------CUCUGGCUCUGGCUCUGUGUCCGACUCUUC .((..(((((((((.......))))))..........(((..((((..((((..(((....)))..))---------------))..))))..)))........)))..)).. ( -29.00, z-score = -0.72, R) >consensus CAGUUUCCAUGGCCCGUUUUUGGCCAUUGAAUGAGUGGCUUUGGCUCCGUGGCCAACUCCACAUCCCC_______________CUCCGUUCCCGAUUCCCGAUCCGUCGUCUA ........((((((.......))))))........(((..(((((......)))))..))).................................................... (-14.09 = -14.12 + 0.03)
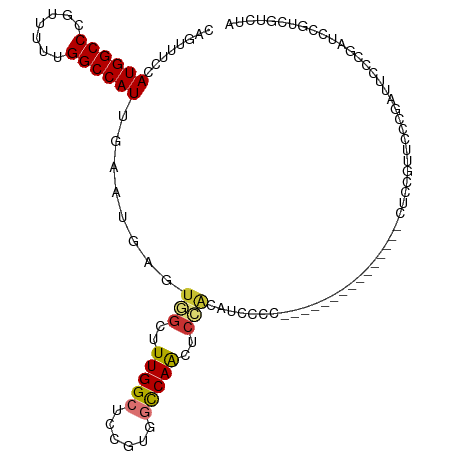
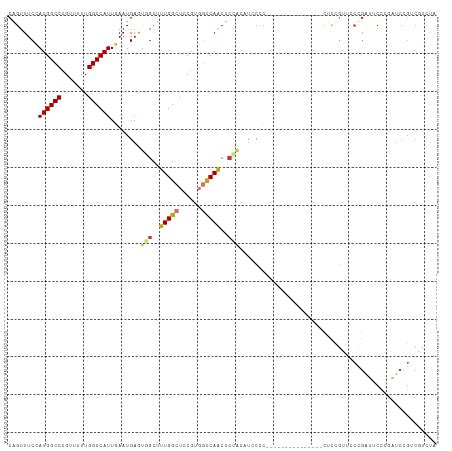
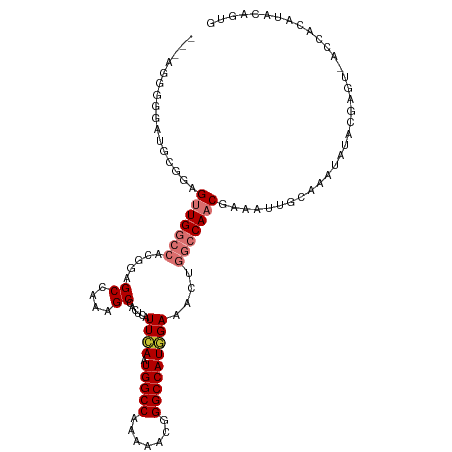
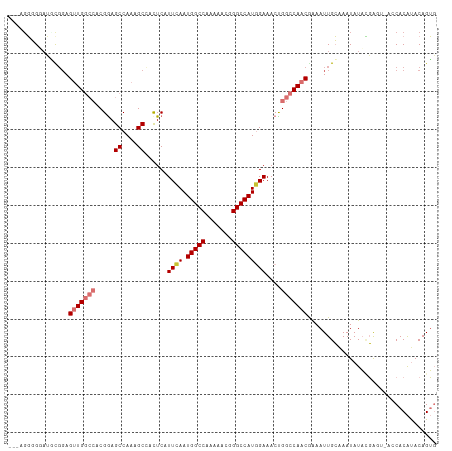
| Location | 25,406,110 – 25,406,220 |
|---|---|
| Length | 110 |
| Sequences | 8 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 81.76 |
| Shannon entropy | 0.34647 |
| G+C content | 0.50402 |
| Mean single sequence MFE | -33.91 |
| Consensus MFE | -19.85 |
| Energy contribution | -20.66 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.601235 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 25406110 110 + 27905053 ---AGGGGUAUGCGGAGUUGGCCACGGAGCCAAAGCCACUCAUUCAAUGGCCAAAAACGGGCCAUGGAAACCGGCCAACGAAAUUGCAAAUAUACGAGUAACCACAUACAGUG ---....(((((.((.(((((((..((........)).....(((.((((((.......)))))).)))...)))))))....((((..........)))))).))))).... ( -35.80, z-score = -2.29, R) >droSim1.chr3R 25078427 110 + 27517382 ---AGGGGUAUGCGGAGUUGGCCACGGAGCCAAAGCCACUCAUUCAAUGGCCAAAAACGGGCCAUGGAAACCGGCCAACGAAAUUGCAAAUAUACGAGUAACCACAUACAGUG ---....(((((.((.(((((((..((........)).....(((.((((((.......)))))).)))...)))))))....((((..........)))))).))))).... ( -35.80, z-score = -2.29, R) >droSec1.super_4 4263751 110 + 6179234 ---AGGGGUAUGCGGAGUUGGCCACGGAGCCAAAGCCACUCAUUCAAUGGCCAAAAACGGGCCAUGGAAACCGGCCAACGAAAUUGCAAAUAUACGAGUAACCACAUACAGUG ---....(((((.((.(((((((..((........)).....(((.((((((.......)))))).)))...)))))))....((((..........)))))).))))).... ( -35.80, z-score = -2.29, R) >droYak2.chr3R 7761078 113 - 28832112 ACAGGGGGGAUUUGGAGUUGGCCACGAAGCCAAAGCCUCUCAUUCAAUGGCCAAAAACGGGCCAUGGAAACUGGCCAACGAAAUUGCAAAUAUACGAGUGACCACAUACAGUG ...((((((.(((((..(((....)))..))))).))))((((((..((........))(((((.(....))))))...................)))))))).......... ( -33.70, z-score = -1.43, R) >droEre2.scaffold_4820 7873712 109 - 10470090 ---AGGGGGCUUUGCAGUUGGCCACGAAGCCAAAGCCACUCAUUCAAUGGCCAAAA-CGGGCCAUGGAAACUGGCCAACGAAAUUGCAAAUAUACGAGUGACCACAUACAGUG ---.((..((((((((((((((((....((....))......(((.((((((....-..)))))).)))..)))))))).....)))).......))))..)).......... ( -33.71, z-score = -1.51, R) >droAna3.scaffold_12911 5101471 105 + 5364042 ---UUGGGGAAACCGAGUUGGCCACGGAGCCAAAGCGAUUCAUUGAAUGGCCAAAAACAGGCCAUGGAAACUGGCCAACAAAAUUGCCAAUAUAUGG-----CACAUACAGUG ---...((....))..((((((((.(...((....(((....))).((((((.......))))))))...))))))))).....(((((.....)))-----))......... ( -36.70, z-score = -3.38, R) >dp4.chr2 2276681 105 + 30794189 ---AGCUGGAGCUGGAGCUGAAGCCGGAGCCAAAGCGAUUCAUUGAAUGGCCAAAAACAGGCCAUCGAAACUGGCCAACAGAAUUGGGAAUAUUGCA-----CACGCACAGGU ---.(((((..((((........))))..)))..(((((((.((((.(((((.......)))))))))..(((.....)))......))..))))).-----...))...... ( -29.90, z-score = -0.58, R) >droPer1.super_7 2441557 105 + 4445127 ---AGCUGGAGCUGGAGCUGAAGCCGGAGCCAAAGCGAUUCAUUGAAUGGCCAAAAACAGGCCAUCGAAACUGGCCAACAGAAUUGGGAAUAUUGCA-----CACGCACAGGU ---.(((((..((((........))))..)))..(((((((.((((.(((((.......)))))))))..(((.....)))......))..))))).-----...))...... ( -29.90, z-score = -0.58, R) >consensus ___AGGGGGAUGCGGAGUUGGCCACGGAGCCAAAGCCACUCAUUCAAUGGCCAAAAACGGGCCAUGGAAACUGGCCAACGAAAUUGCAAAUAUACGAGU_ACCACAUACAGUG ................(((((((.....((....))......((((.(((((.......)))))))))....))))))).................................. (-19.85 = -20.66 + 0.81)
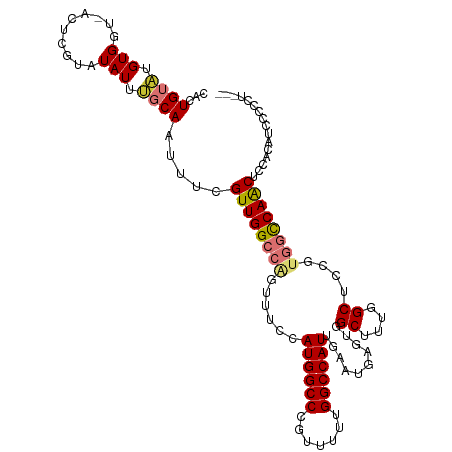
| Location | 25,406,110 – 25,406,220 |
|---|---|
| Length | 110 |
| Sequences | 8 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 81.76 |
| Shannon entropy | 0.34647 |
| G+C content | 0.50402 |
| Mean single sequence MFE | -34.57 |
| Consensus MFE | -21.09 |
| Energy contribution | -22.00 |
| Covariance contribution | 0.91 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.596666 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
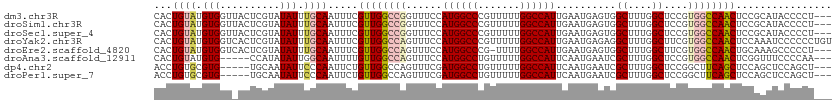
>dm3.chr3R 25406110 110 - 27905053 CACUGUAUGUGGUUACUCGUAUAUUUGCAAUUUCGUUGGCCGGUUUCCAUGGCCCGUUUUUGGCCAUUGAAUGAGUGGCUUUGGCUCCGUGGCCAACUCCGCAUACCCCU--- ....((((((((......(((....)))......((((((((..(((.((((((.......)))))).)))...(..((....))..).)))))))).))))))))....--- ( -39.50, z-score = -2.88, R) >droSim1.chr3R 25078427 110 - 27517382 CACUGUAUGUGGUUACUCGUAUAUUUGCAAUUUCGUUGGCCGGUUUCCAUGGCCCGUUUUUGGCCAUUGAAUGAGUGGCUUUGGCUCCGUGGCCAACUCCGCAUACCCCU--- ....((((((((......(((....)))......((((((((..(((.((((((.......)))))).)))...(..((....))..).)))))))).))))))))....--- ( -39.50, z-score = -2.88, R) >droSec1.super_4 4263751 110 - 6179234 CACUGUAUGUGGUUACUCGUAUAUUUGCAAUUUCGUUGGCCGGUUUCCAUGGCCCGUUUUUGGCCAUUGAAUGAGUGGCUUUGGCUCCGUGGCCAACUCCGCAUACCCCU--- ....((((((((......(((....)))......((((((((..(((.((((((.......)))))).)))...(..((....))..).)))))))).))))))))....--- ( -39.50, z-score = -2.88, R) >droYak2.chr3R 7761078 113 + 28832112 CACUGUAUGUGGUCACUCGUAUAUUUGCAAUUUCGUUGGCCAGUUUCCAUGGCCCGUUUUUGGCCAUUGAAUGAGAGGCUUUGGCUUCGUGGCCAACUCCAAAUCCCCCCUGU ...((((((.(....).))))))...........((((((((..(((.((((((.......)))))).)))...(((((....))))).))))))))................ ( -35.70, z-score = -2.09, R) >droEre2.scaffold_4820 7873712 109 + 10470090 CACUGUAUGUGGUCACUCGUAUAUUUGCAAUUUCGUUGGCCAGUUUCCAUGGCCCG-UUUUGGCCAUUGAAUGAGUGGCUUUGGCUUCGUGGCCAACUGCAAAGCCCCCU--- ...((((((.(....).))))))((((((.....((((((((..(((.((((((..-....)))))).)))...(.(((....))).).)))))))))))))).......--- ( -36.10, z-score = -1.67, R) >droAna3.scaffold_12911 5101471 105 - 5364042 CACUGUAUGUG-----CCAUAUAUUGGCAAUUUUGUUGGCCAGUUUCCAUGGCCUGUUUUUGGCCAUUCAAUGAAUCGCUUUGGCUCCGUGGCCAACUCGGUUUCCCCAA--- .((((....((-----(((.....))))).....((((((((......((((((.......))))))..........((....))....)))))))).))))........--- ( -33.30, z-score = -2.01, R) >dp4.chr2 2276681 105 - 30794189 ACCUGUGCGUG-----UGCAAUAUUCCCAAUUCUGUUGGCCAGUUUCGAUGGCCUGUUUUUGGCCAUUCAAUGAAUCGCUUUGGCUCCGGCUUCAGCUCCAGCUCCAGCU--- ..((((((...-----.)))......(((((...))))).)))....(((((((.......))))))).........(((..((((..(((....)))..))))..))).--- ( -26.50, z-score = 0.36, R) >droPer1.super_7 2441557 105 - 4445127 ACCUGUGCGUG-----UGCAAUAUUCCCAAUUCUGUUGGCCAGUUUCGAUGGCCUGUUUUUGGCCAUUCAAUGAAUCGCUUUGGCUCCGGCUUCAGCUCCAGCUCCAGCU--- ..((((((...-----.)))......(((((...))))).)))....(((((((.......))))))).........(((..((((..(((....)))..))))..))).--- ( -26.50, z-score = 0.36, R) >consensus CACUGUAUGUGGU_ACUCGUAUAUUUGCAAUUUCGUUGGCCAGUUUCCAUGGCCCGUUUUUGGCCAUUGAAUGAGUGGCUUUGGCUCCGUGGCCAACUCCACAUCCCCCU___ ...((((((........))))))...........((((((((......((((((.......))))))..........((....))....))))))))................ (-21.09 = -22.00 + 0.91)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:55:07 2011