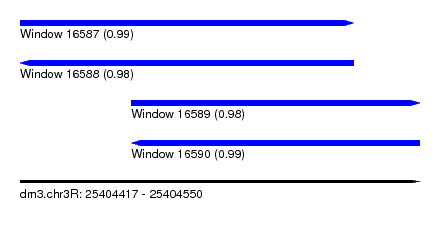
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 25,404,417 – 25,404,550 |
| Length | 133 |
| Max. P | 0.991100 |

| Location | 25,404,417 – 25,404,528 |
|---|---|
| Length | 111 |
| Sequences | 9 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 72.99 |
| Shannon entropy | 0.56234 |
| G+C content | 0.45856 |
| Mean single sequence MFE | -30.83 |
| Consensus MFE | -17.37 |
| Energy contribution | -17.37 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.45 |
| SVM RNA-class probability | 0.990927 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
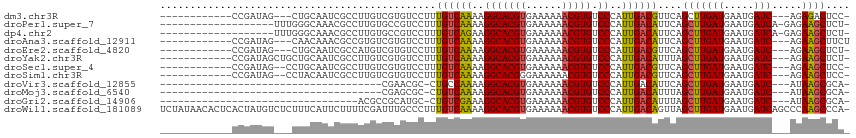
>dm3.chr3R 25404417 111 + 27905053 GAAAAAGGCCAAAUACU--GGAAUGCCGAGUGUAAGCCAGGA-GUCUCUGAUCAUUCAUCAAGCUGAACGUCAAUGGGACACGUUUUUUCACGUGCCUUUUGACAAAGGACACG (((((((((...(((((--((....)).)))))..)))....-(((((((((..((((......)))).))))..)))))....)))))).(((((((((....))))).)))) ( -31.00, z-score = -1.39, R) >droPer1.super_7 2439817 105 + 4445127 ---GGUCCAUGUCCAGUCUCUGGGACUCUGUUAGAGCUUCU------CUGAUCAUUCAUCAAGCUGAAUGUCAAUGGGACACGUUUUUUCACGUGCCUUUUGACAAAGGACGGC ---.((((..((((........))))........(((((..------.(((....)))..)))))...((((((.(((.(((((......)))))))).))))))..))))... ( -34.00, z-score = -2.09, R) >dp4.chr2 2274964 105 + 30794189 ---GGUCCAUGUCCAGUCUCUGGGACUCUGUUAGAGCUUCU------CUGAUCAUUCAUCAAGCUGAAUGUCAAUGGGACACGUUUUUUCACGUGCCUUCUGACAAAGGACGGC ---.((((..((((........))))..(((((((......------.(((.((((((......)))))))))..(((.(((((......)))))))))))))))..))))... ( -34.40, z-score = -2.06, R) >droAna3.scaffold_12911 5099629 113 + 5364042 -GAAAGAAUAAUAUACUCAAAGAGACCCAGCCGAACCCAAGAAGCUUCUGAUCAUUCAUCAAGCUGAAUGUCAAUGGGACACGUUUUUUCACGUGCCUUUUGACAAAGGACACG -((((((((......(((...))).((((...((........(((((.(((....)))..))))).....))..))))....)))))))).(((((((((....))))).)))) ( -27.32, z-score = -2.24, R) >droEre2.scaffold_4820 7872006 92 - 10470090 ---------------GA--AAAAGGCCAAAUA----CUGAGA-GCUUCUGAUCAUUCAUCAAGCUGAACGUCAAUGGGACACGUUUUUUCACGUGCCUUUUGACAAAGGACACG ---------------..--....(.((.....----.....(-((((.(((....)))..)))))....(((((.(((.(((((......)))))))).)))))...)).)... ( -24.30, z-score = -2.01, R) >droYak2.chr3R 7759245 111 - 28832112 GAAAAAGGCCAAAUACU--GGAGUGCCGAGUGUAAGCCAAGA-GCUUCUGAUCAUUCAUCAAGCUAAAUGUCAAUGGGACACGUUUUUUCACGUGCCUUUUGACAAAGGACACG ......(((...(((((--((....)).)))))..)))...(-((((.(((....)))..)))))...((((((.(((.(((((......)))))))).))))))......... ( -32.50, z-score = -2.24, R) >droSec1.super_4 4262065 110 + 6179234 -GAAACGGCCAAAUACU--GGAAUGCCGAGUGUAACCCAGGA-GCUUCUGAUCAUUCAUCAAGCUGAACGUCAAUGGGACACGUUUUUUCACGUGCCUUUUGACAAAGGACACG -....((((........--.....)))).((((...((...(-((((.(((....)))..)))))....(((((.(((.(((((......)))))))).)))))...)))))). ( -32.22, z-score = -2.03, R) >droSim1.chr3R 25076723 110 + 27517382 -GAAAAGGCCAAAUACU--GGAAUGCCGAGUGUAACCCAGGA-GCUUCUGAUCAUUCAUCAAGCUGAACGUCAAUGGGACACGUUUUUUCCCGUGCCUUUUGACAAAGGACACG -((((((((...(((((--((....)).)))))..((((..(-((((.(((....)))..)))))(.....)..))))....)))))))).(((((((((....))))).)))) ( -29.00, z-score = -0.94, R) >droWil1.scaffold_181089 5668981 98 + 12369635 -----------GAUGGGCCAAAUGACUUUGCUUGGGCUUGGG-----CUGAUCAUUCAUCAAGCUAACUGUCAAUGGGACACGUUUUUUCACGUGCCUUUUGACAAAGGGCAAA -----------(((.((((......((......)).....))-----)).))).........(((...((((((.(((.(((((......)))))))).))))))...)))... ( -32.70, z-score = -2.80, R) >consensus __AAAAGGCCAAAUACU__GGAAGGCCGAGUGUAAGCCAGGA_GCUUCUGAUCAUUCAUCAAGCUGAAUGUCAAUGGGACACGUUUUUUCACGUGCCUUUUGACAAAGGACACG .........................((.....................((((.....))))........(((((.(((.(((((......)))))))).)))))...))..... (-17.37 = -17.37 + -0.00)

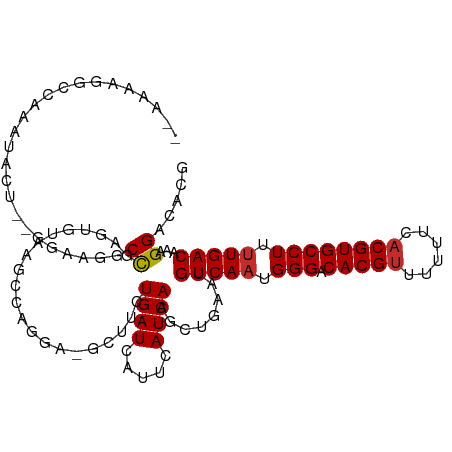
| Location | 25,404,417 – 25,404,528 |
|---|---|
| Length | 111 |
| Sequences | 9 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 72.99 |
| Shannon entropy | 0.56234 |
| G+C content | 0.45856 |
| Mean single sequence MFE | -29.39 |
| Consensus MFE | -17.49 |
| Energy contribution | -17.16 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.981275 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 25404417 111 - 27905053 CGUGUCCUUUGUCAAAAGGCACGUGAAAAAACGUGUCCCAUUGACGUUCAGCUUGAUGAAUGAUCAGAGAC-UCCUGGCUUACACUCGGCAUUCC--AGUAUUUGGCCUUUUUC ((((.(((((....))))))))).((((((..(.(((...((((((((((......)))))).)))).)))-.)..((((...(((.((....))--)))....)))))))))) ( -29.10, z-score = -1.06, R) >droPer1.super_7 2439817 105 - 4445127 GCCGUCCUUUGUCAAAAGGCACGUGAAAAAACGUGUCCCAUUGACAUUCAGCUUGAUGAAUGAUCAG------AGAAGCUCUAACAGAGUCCCAGAGACUGGACAUGGACC--- ...((((..((((....(((((((......)))))))...((((((((((......)))))).))))------.(..((((.....))))..)........)))).)))).--- ( -32.30, z-score = -2.67, R) >dp4.chr2 2274964 105 - 30794189 GCCGUCCUUUGUCAGAAGGCACGUGAAAAAACGUGUCCCAUUGACAUUCAGCUUGAUGAAUGAUCAG------AGAAGCUCUAACAGAGUCCCAGAGACUGGACAUGGACC--- ...(((((((((.(((.(((((((......)))))))...((((((((((......)))))).))))------......))).)))))((((........))))..)))).--- ( -34.10, z-score = -2.91, R) >droAna3.scaffold_12911 5099629 113 - 5364042 CGUGUCCUUUGUCAAAAGGCACGUGAAAAAACGUGUCCCAUUGACAUUCAGCUUGAUGAAUGAUCAGAAGCUUCUUGGGUUCGGCUGGGUCUCUUUGAGUAUAUUAUUCUUUC- .(.(.((..((((((..(((((((......))))).))..))))))..(((((((((.....))))(((.((....)).)))))))))).).)...(((((...)))))....- ( -30.10, z-score = -1.32, R) >droEre2.scaffold_4820 7872006 92 + 10470090 CGUGUCCUUUGUCAAAAGGCACGUGAAAAAACGUGUCCCAUUGACGUUCAGCUUGAUGAAUGAUCAGAAGC-UCUCAG----UAUUUGGCCUUUU--UC--------------- ..........((((((.(((((((......)))))))...((((((((((......)))))).))))....-......----..)))))).....--..--------------- ( -22.60, z-score = -1.16, R) >droYak2.chr3R 7759245 111 + 28832112 CGUGUCCUUUGUCAAAAGGCACGUGAAAAAACGUGUCCCAUUGACAUUUAGCUUGAUGAAUGAUCAGAAGC-UCUUGGCUUACACUCGGCACUCC--AGUAUUUGGCCUUUUUC .((((((..((((((..(((((((......))))).))..))))))...(((((..(((....))).))))-)...))...))))..(((.(...--.......))))...... ( -28.50, z-score = -1.17, R) >droSec1.super_4 4262065 110 - 6179234 CGUGUCCUUUGUCAAAAGGCACGUGAAAAAACGUGUCCCAUUGACGUUCAGCUUGAUGAAUGAUCAGAAGC-UCCUGGGUUACACUCGGCAUUCC--AGUAUUUGGCCGUUUC- .(((((((..(((((..(((((((......))))).))..)))))....(((((..(((....))).))))-)...)))..)))).((((.....--........))))....- ( -30.22, z-score = -1.02, R) >droSim1.chr3R 25076723 110 - 27517382 CGUGUCCUUUGUCAAAAGGCACGGGAAAAAACGUGUCCCAUUGACGUUCAGCUUGAUGAAUGAUCAGAAGC-UCCUGGGUUACACUCGGCAUUCC--AGUAUUUGGCCUUUUC- ((((.(((((....))))))))).(((((...((((((((((((((((((......)))))).))))....-...))))..))))..(((.....--........))))))))- ( -29.53, z-score = -0.57, R) >droWil1.scaffold_181089 5668981 98 - 12369635 UUUGCCCUUUGUCAAAAGGCACGUGAAAAAACGUGUCCCAUUGACAGUUAGCUUGAUGAAUGAUCAG-----CCCAAGCCCAAGCAAAGUCAUUUGGCCCAUC----------- (((((...(((((((..(((((((......))))).))..)))))))...(((((.(((....))).-----..)))))....)))))(((....))).....----------- ( -28.10, z-score = -3.06, R) >consensus CGUGUCCUUUGUCAAAAGGCACGUGAAAAAACGUGUCCCAUUGACAUUCAGCUUGAUGAAUGAUCAGAAGC_UCCUGGCUUACACUCGGCCUUCC__AGUAUUUGGCCUUUU__ ...((....((((((..(((((((......))))).))..))))))....))(((((.....)))))............................................... (-17.49 = -17.16 + -0.33)
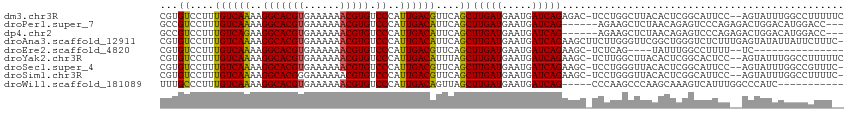
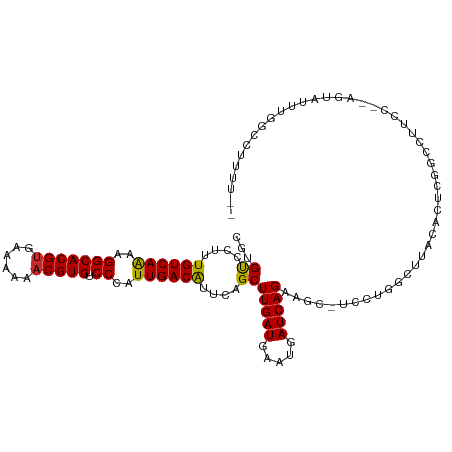
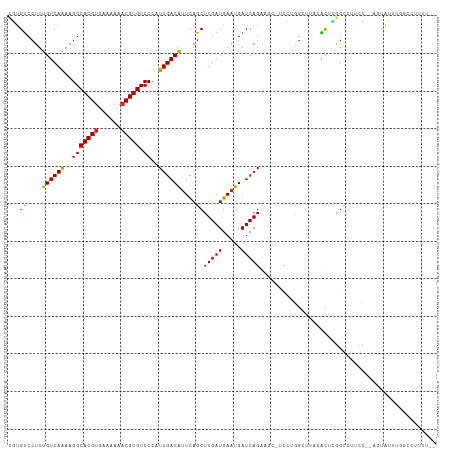
| Location | 25,404,454 – 25,404,550 |
|---|---|
| Length | 96 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 74.95 |
| Shannon entropy | 0.48244 |
| G+C content | 0.46236 |
| Mean single sequence MFE | -26.86 |
| Consensus MFE | -16.93 |
| Energy contribution | -17.12 |
| Covariance contribution | 0.18 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.979890 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 25404454 96 + 27905053 -GGAGUCUCU---GAUCAUUCAUCAAGCUGAACGUCAAUGGGACACGUUUUUUCACGUGCCUUUUGACAAAGGACACGACAAGGCGAUUGCAG---CUAUCGG------------ -...(((.((---.....((((......)))).(((((.(((.(((((......)))))))).)))))..))))).(((...(((.......)---)).))).------------ ( -25.40, z-score = -1.00, R) >droPer1.super_7 2439846 94 + 4445127 -AGAGCUUCUC-UGAUCAUUCAUCAAGCUGAAUGUCAAUGGGACACGUUUUUUCACGUGCCUUUUGACAAAGGACGGCACAAGGCGUUUGCCCAAA------------------- -..(((((...-(((....)))..)))))...((((((.(((.(((((......)))))))).))))))..(((((.(....).))))).......------------------- ( -27.50, z-score = -1.57, R) >dp4.chr2 2274993 94 + 30794189 -AGAGCUUCUC-UGAUCAUUCAUCAAGCUGAAUGUCAAUGGGACACGUUUUUUCACGUGCCUUCUGACAAAGGACGGCACAAGGCGUUUGCCCAAA------------------- -((((...)))-)((.((((((......))))))))..((((..(((((((.....(((((((((.....)))).))))))))))))...))))..------------------- ( -29.10, z-score = -1.98, R) >droAna3.scaffold_12911 5099667 97 + 5364042 AGAAGCUUCU---GAUCAUUCAUCAAGCUGAAUGUCAAUGGGACACGUUUUUUCACGUGCCUUUUGACAAAGGACACGACACGGCGUUUGUUG---CUAUCGG------------ ...(((((.(---((....)))..)))))...((((((.(((.(((((......)))))))).)))))).......(((...((((.....))---)).))).------------ ( -27.70, z-score = -1.68, R) >droEre2.scaffold_4820 7872024 96 - 10470090 -AGAGCUUCU---GAUCAUUCAUCAAGCUGAACGUCAAUGGGACACGUUUUUUCACGUGCCUUUUGACAAAGGACACGACAUGGCGAUUGCAG---CUAUCGG------------ -..(((((.(---((....)))..)))))....(((((.(((.(((((......)))))))).)))))........(((..((((.......)---)))))).------------ ( -27.80, z-score = -1.97, R) >droYak2.chr3R 7759282 99 - 28832112 -AGAGCUUCU---GAUCAUUCAUCAAGCUAAAUGUCAAUGGGACACGUUUUUUCACGUGCCUUUUGACAAAGGACACGACAAGGCGAUUGCAGCAGCUAUCGG------------ -..(((((.(---((....)))..)))))...((((((.(((.(((((......)))))))).))))))...............((((.((....)).)))).------------ ( -28.50, z-score = -2.14, R) >droSec1.super_4 4262101 97 + 6179234 -GGAGCUUCU---GAUCAUUCAUCAAGCUGAACGUCAAUGGGACACGUUUUUUCACGUGCCUUUUGACAAAGGACACGACAAGGCGAUUGCAGG--CUAUCGG------------ -(((((((.(---((((.((((......)))).(((((.(((.(((((......)))))))).))))).................))))).)))--)).))..------------ ( -26.20, z-score = -1.30, R) >droSim1.chr3R 25076759 97 + 27517382 -GGAGCUUCU---GAUCAUUCAUCAAGCUGAACGUCAAUGGGACACGUUUUUUCCCGUGCCUUUUGACAAAGGACACGACAAGGCGAUUGUAGG--CUAUCGG------------ -(((((((..---((((.((((......)))).(((...((((.........))))((((((((....))))).)))))).....))))..)))--)).))..------------ ( -25.90, z-score = -0.80, R) >droVir3.scaffold_12855 743645 73 + 10161210 -UGCGCUUAU---GAUCAUUCAUCAAGCUGAAUGUCAAUGGGACACGUUUUUUCACGUGCCUUUUGGCAG-GCGUUCG------------------------------------- -.((((((((---((....)))).))))....((((((.(((.(((((......)))))))).)))))).-)).....------------------------------------- ( -25.30, z-score = -3.24, R) >droMoj3.scaffold_6540 21922282 73 - 34148556 -UGCGCUUAU---GAUCAUUCAUCAAGCUAAAUGUCAAUGGGACACGUUUUUUCACGUGCCUUUUGACAG-GCGCUCG------------------------------------- -.((((((((---((....))))..........(((((.(((.(((((......)))))))).)))))))-))))...------------------------------------- ( -26.10, z-score = -4.30, R) >droGri2.scaffold_14906 1849435 77 + 14172833 -UGCGCUUAU---GAUCAUUCAUCAAGCUAAAUGUCAAUGGGACACGUUUUUUCACGUGCCUUUCGACAG-GCAUGCGGCGU--------------------------------- -(((((((((---((....)))).))))....((((...(((.(((((......))))))))...)))).-)))........--------------------------------- ( -23.10, z-score = -1.82, R) >droWil1.scaffold_181089 5669002 114 + 12369635 -UGGGCUUGGGCUGAUCAUUCAUCAAGCUAACUGUCAAUGGGACACGUUUUUUCACGUGCCUUUUGACAAAGGGCAAAUCGAAAAGAAUGAAAGAGACAUAGUGAGUGUUAUAGA -..(((....)))..((.(((((...(((...((((((.(((.(((((......)))))))).))))))...)))...((.....))))))).))(((((.....)))))..... ( -29.70, z-score = -1.22, R) >consensus _AGAGCUUCU___GAUCAUUCAUCAAGCUGAAUGUCAAUGGGACACGUUUUUUCACGUGCCUUUUGACAAAGGACACGACAAGGCGAUUGCAG___CUAUCGG____________ ....((((.....((....))...)))).....(((((.(((.(((((......)))))))).)))))............................................... (-16.93 = -17.12 + 0.18)

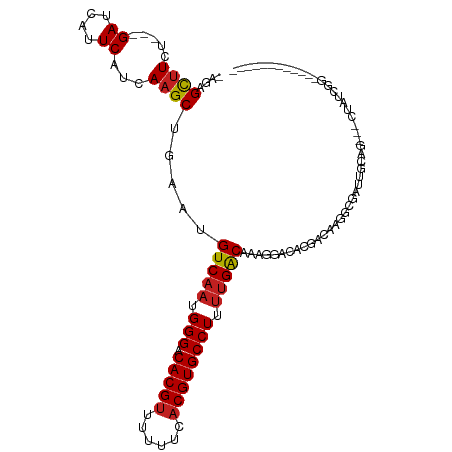
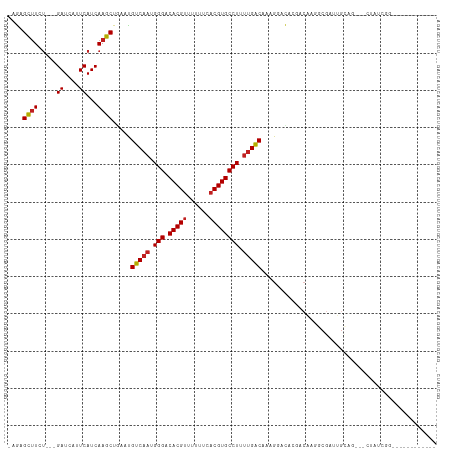
| Location | 25,404,454 – 25,404,550 |
|---|---|
| Length | 96 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 74.95 |
| Shannon entropy | 0.48244 |
| G+C content | 0.46236 |
| Mean single sequence MFE | -25.54 |
| Consensus MFE | -16.71 |
| Energy contribution | -16.37 |
| Covariance contribution | -0.34 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.46 |
| SVM RNA-class probability | 0.991100 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
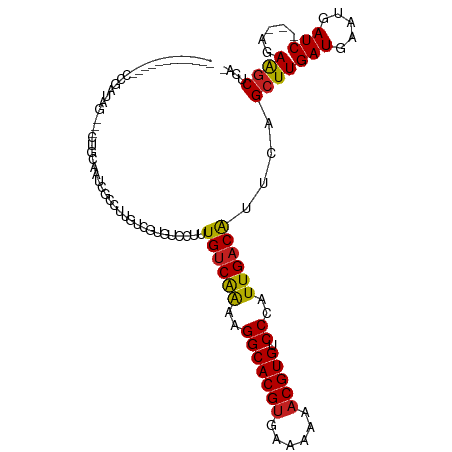
>dm3.chr3R 25404454 96 - 27905053 ------------CCGAUAG---CUGCAAUCGCCUUGUCGUGUCCUUUGUCAAAAGGCACGUGAAAAAACGUGUCCCAUUGACGUUCAGCUUGAUGAAUGAUC---AGAGACUCC- ------------.((((((---..((....)).)))))).(((.((((((((..(((((((......))))).))..))))((((((......))))))..)---))))))...- ( -26.20, z-score = -1.89, R) >droPer1.super_7 2439846 94 - 4445127 -------------------UUUGGGCAAACGCCUUGUGCCGUCCUUUGUCAAAAGGCACGUGAAAAAACGUGUCCCAUUGACAUUCAGCUUGAUGAAUGAUCA-GAGAAGCUCU- -------------------...((((..(((.(....).))).(((((((((..(((((((......))))).))..))))((((((......))))))..))-)))..)))).- ( -27.00, z-score = -1.66, R) >dp4.chr2 2274993 94 - 30794189 -------------------UUUGGGCAAACGCCUUGUGCCGUCCUUUGUCAGAAGGCACGUGAAAAAACGUGUCCCAUUGACAUUCAGCUUGAUGAAUGAUCA-GAGAAGCUCU- -------------------..(((((....(((((.((.((.....)).)).)))))((((......))))).))))((((((((((......)))))).)))-).........- ( -28.10, z-score = -1.72, R) >droAna3.scaffold_12911 5099667 97 - 5364042 ------------CCGAUAG---CAACAAACGCCGUGUCGUGUCCUUUGUCAAAAGGCACGUGAAAAAACGUGUCCCAUUGACAUUCAGCUUGAUGAAUGAUC---AGAAGCUUCU ------------.((((((---(.......))..))))).......((((((..(((((((......))))).))..))))))...(((((..(((....))---).)))))... ( -25.00, z-score = -1.59, R) >droEre2.scaffold_4820 7872024 96 + 10470090 ------------CCGAUAG---CUGCAAUCGCCAUGUCGUGUCCUUUGUCAAAAGGCACGUGAAAAAACGUGUCCCAUUGACGUUCAGCUUGAUGAAUGAUC---AGAAGCUCU- ------------.(((((.---..((....))..)))))........(((((..(((((((......))))).))..)))))....(((((..(((....))---).)))))..- ( -26.00, z-score = -1.73, R) >droYak2.chr3R 7759282 99 + 28832112 ------------CCGAUAGCUGCUGCAAUCGCCUUGUCGUGUCCUUUGUCAAAAGGCACGUGAAAAAACGUGUCCCAUUGACAUUUAGCUUGAUGAAUGAUC---AGAAGCUCU- ------------.....((((.(((..((((..(..(((.((....((((((..(((((((......))))).))..))))))....)).)))..).)))))---)).))))..- ( -29.20, z-score = -2.77, R) >droSec1.super_4 4262101 97 - 6179234 ------------CCGAUAG--CCUGCAAUCGCCUUGUCGUGUCCUUUGUCAAAAGGCACGUGAAAAAACGUGUCCCAUUGACGUUCAGCUUGAUGAAUGAUC---AGAAGCUCC- ------------.((((((--...((....)).))))))........(((((..(((((((......))))).))..)))))....(((((..(((....))---).)))))..- ( -25.10, z-score = -1.80, R) >droSim1.chr3R 25076759 97 - 27517382 ------------CCGAUAG--CCUACAAUCGCCUUGUCGUGUCCUUUGUCAAAAGGCACGGGAAAAAACGUGUCCCAUUGACGUUCAGCUUGAUGAAUGAUC---AGAAGCUCC- ------------..(((((--(........))..))))(((.(((((....))))))))((((.........)))).((((((((((......)))))).))---)).......- ( -24.00, z-score = -0.95, R) >droVir3.scaffold_12855 743645 73 - 10161210 -------------------------------------CGAACGC-CUGCCAAAAGGCACGUGAAAAAACGUGUCCCAUUGACAUUCAGCUUGAUGAAUGAUC---AUAAGCGCA- -------------------------------------....(((-.((((....)))).))).......((((.....(((((((((......)))))).))---)...)))).- ( -20.70, z-score = -2.48, R) >droMoj3.scaffold_6540 21922282 73 + 34148556 -------------------------------------CGAGCGC-CUGUCAAAAGGCACGUGAAAAAACGUGUCCCAUUGACAUUUAGCUUGAUGAAUGAUC---AUAAGCGCA- -------------------------------------...((..-.((((((..(((((((......))))).))..))))))....(((((.(((....))---)))))))).- ( -24.60, z-score = -3.63, R) >droGri2.scaffold_14906 1849435 77 - 14172833 ---------------------------------ACGCCGCAUGC-CUGUCGAAAGGCACGUGAAAAAACGUGUCCCAUUGACAUUUAGCUUGAUGAAUGAUC---AUAAGCGCA- ---------------------------------.....((....-.((((((..(((((((......))))).))..))))))....(((((.(((....))---)))))))).- ( -24.40, z-score = -2.66, R) >droWil1.scaffold_181089 5669002 114 - 12369635 UCUAUAACACUCACUAUGUCUCUUUCAUUCUUUUCGAUUUGCCCUUUGUCAAAAGGCACGUGAAAAAACGUGUCCCAUUGACAGUUAGCUUGAUGAAUGAUCAGCCCAAGCCCA- ........................((((((...((((...((...(((((((..(((((((......))))).))..)))))))...)))))).))))))..............- ( -26.20, z-score = -2.51, R) >consensus ____________CCGAUAG___CUGCAAUCGCCUUGUCGUGUCCUUUGUCAAAAGGCACGUGAAAAAACGUGUCCCAUUGACAUUCAGCUUGAUGAAUGAUC___AGAAGCUCA_ ...............................................(((((..(((((((......))))).))..)))))(((((......)))))................. (-16.71 = -16.37 + -0.34)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:55:04 2011