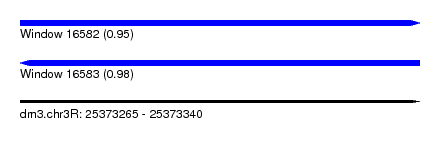
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 25,373,265 – 25,373,340 |
| Length | 75 |
| Max. P | 0.975391 |

| Location | 25,373,265 – 25,373,340 |
|---|---|
| Length | 75 |
| Sequences | 12 |
| Columns | 77 |
| Reading direction | forward |
| Mean pairwise identity | 94.13 |
| Shannon entropy | 0.12846 |
| G+C content | 0.38090 |
| Mean single sequence MFE | -18.57 |
| Consensus MFE | -16.64 |
| Energy contribution | -16.73 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.952499 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
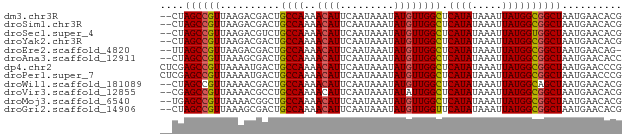
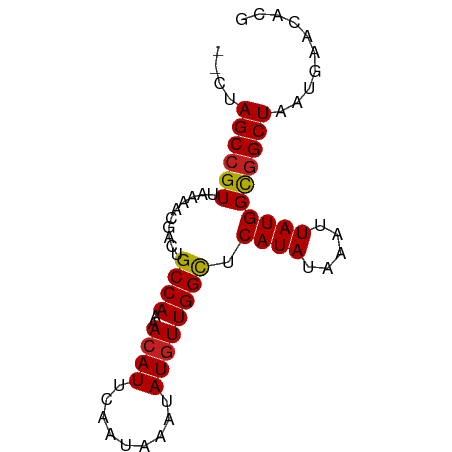
>dm3.chr3R 25373265 75 + 27905053 CGUGUUCAUUAGCCGCCAUAAUUUAUAUGAGCCAACAUAUUUAUUGAAUGUUUUGGCAGUCGUCUUAACGGCUAG-- .........((((((.((((.....)))).(((((.((((((...)))))).)))))...........)))))).-- ( -19.30, z-score = -2.53, R) >droSim1.chr3R 25046455 75 + 27517382 CGUGUUCAUUAGCCGCCAUAAUUUAUAUGAGCCAACAUAUUUAUUGAAUGUUUUGGCAGUCGUCUUAACGGCUAG-- .........((((((.((((.....)))).(((((.((((((...)))))).)))))...........)))))).-- ( -19.30, z-score = -2.53, R) >droSec1.super_4 4231504 75 + 6179234 CGUGUUCAUUAGCCACCAUAAUUUAUAUGAGCCAACAUAUUUAUUGAAUGUUUUGGCAGACGUCUUAACGGCUAG-- .........(((((..((((.....)))).(((((.((((((...)))))).)))))............))))).-- ( -18.00, z-score = -2.50, R) >droYak2.chr3R 7726613 75 - 28832112 CGUGUUCAUUAGCCGCCAUAAUUUAUAUGAGCCAACAUAUUUAUUGAAUGUUUUGGCAGUCGUCUUAACGGCUAG-- .........((((((.((((.....)))).(((((.((((((...)))))).)))))...........)))))).-- ( -19.30, z-score = -2.53, R) >droEre2.scaffold_4820 7841322 74 - 10470090 -CUGUUCAUUAGCCGCCAUAAUUUAUAUGAGCCAACAUAUUUAUUGAAUGUUUUGGCAGUCGUCUUAACGGCUAA-- -.......(((((((.((((.....)))).(((((.((((((...)))))).)))))...........)))))))-- ( -19.50, z-score = -2.90, R) >droAna3.scaffold_12911 5067740 75 + 5364042 GGUGUUCAUUAGCCGCCAUAAUUUAUAUGAGCCAACAUAUUUAUUGAAUGUUUUGGCAGUCGCUUUAACGGCUAG-- .........((((((.((((.....)))).(((((.((((((...)))))).)))))...........)))))).-- ( -19.30, z-score = -1.98, R) >dp4.chr2 2241511 77 + 30794189 CGGGUUCAUUAGCCGCCAUAAUUUAUAUGAGCCAACAUAUUUAUUGAAUGUUUUGGCAGUCAUUUUAACGGCUCGAG ..........(((((.((((.....)))).(((((.((((((...)))))).)))))...........))))).... ( -17.30, z-score = -1.51, R) >droPer1.super_7 2406338 77 + 4445127 CGGGUUCAUUAGCCGCCAUAAUUUAUAUGAGCCAACAUAUUUAUUGAAUGUUUUGGCAGUCAUUUUAACGGCUCGAG ..........(((((.((((.....)))).(((((.((((((...)))))).)))))...........))))).... ( -17.30, z-score = -1.51, R) >droWil1.scaffold_181089 5624781 75 + 12369635 CGUGUUCAUUAGCUGCCAUAAUUUAUAUGAGCCAACAUAUUUAUUGAAUGUUUUGGCAGUCGUUUUAACGGCUAG-- (((((..((((.......))))..))))).(((((.((((((...)))))).)))))((((((....))))))..-- ( -18.20, z-score = -2.07, R) >droVir3.scaffold_12855 705520 75 + 10161210 CGUGUUCAUUAGCCGCCAUAAUUUAUAUGAGCCAAUAUAUUUAUUGAAUGUUUUGGCAGGCGUUUUAACGGCUCG-- ..........((((((((((.....)))..(((((.((((((...)))))).))))).)))((....))))))..-- ( -18.10, z-score = -1.43, R) >droMoj3.scaffold_6540 21884923 75 - 34148556 CGUGUUCAUUAGCCGCCAUAAUUUAUAUGAGCCAACAUAUUUAUUGAAUGUUUUGGCAGCCGUUUUAACGGCUCA-- (((((..((((.......))))..))))).(((((.((((((...)))))).)))))((((((....))))))..-- ( -20.30, z-score = -2.70, R) >droGri2.scaffold_14906 1809573 75 + 14172833 CGUGUUCAUUAGCCGCCAUAAUUUAUAUGAACCAACAUAUUUAUUGAAUGUUUUGGCAGUCGCUUUAACGGCUAG-- .........(((((....((((..(((((......)))))..))))...(((..(((....)))..)))))))).-- ( -16.90, z-score = -1.83, R) >consensus CGUGUUCAUUAGCCGCCAUAAUUUAUAUGAGCCAACAUAUUUAUUGAAUGUUUUGGCAGUCGUUUUAACGGCUAG__ ..........(((((.((((.....)))).(((((.((((((...)))))).)))))...........))))).... (-16.64 = -16.73 + 0.09)

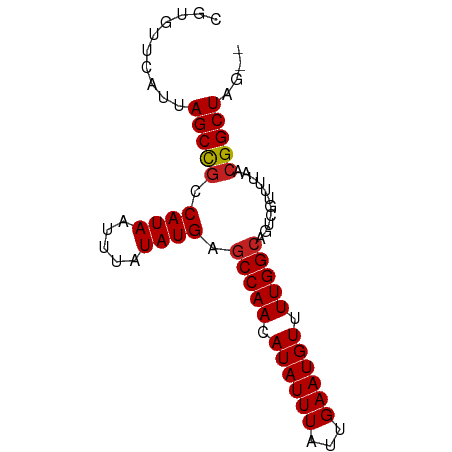
| Location | 25,373,265 – 25,373,340 |
|---|---|
| Length | 75 |
| Sequences | 12 |
| Columns | 77 |
| Reading direction | reverse |
| Mean pairwise identity | 94.13 |
| Shannon entropy | 0.12846 |
| G+C content | 0.38090 |
| Mean single sequence MFE | -17.15 |
| Consensus MFE | -15.53 |
| Energy contribution | -15.54 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.975391 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 25373265 75 - 27905053 --CUAGCCGUUAAGACGACUGCCAAAACAUUCAAUAAAUAUGUUGGCUCAUAUAAAUUAUGGCGGCUAAUGAACACG --.(((((((..........((((..((((.........)))))))).((((.....)))))))))))......... ( -18.20, z-score = -2.89, R) >droSim1.chr3R 25046455 75 - 27517382 --CUAGCCGUUAAGACGACUGCCAAAACAUUCAAUAAAUAUGUUGGCUCAUAUAAAUUAUGGCGGCUAAUGAACACG --.(((((((..........((((..((((.........)))))))).((((.....)))))))))))......... ( -18.20, z-score = -2.89, R) >droSec1.super_4 4231504 75 - 6179234 --CUAGCCGUUAAGACGUCUGCCAAAACAUUCAAUAAAUAUGUUGGCUCAUAUAAAUUAUGGUGGCUAAUGAACACG --.((((((...........((((..((((.........))))))))(((((.....)))))))))))......... ( -15.30, z-score = -1.52, R) >droYak2.chr3R 7726613 75 + 28832112 --CUAGCCGUUAAGACGACUGCCAAAACAUUCAAUAAAUAUGUUGGCUCAUAUAAAUUAUGGCGGCUAAUGAACACG --.(((((((..........((((..((((.........)))))))).((((.....)))))))))))......... ( -18.20, z-score = -2.89, R) >droEre2.scaffold_4820 7841322 74 + 10470090 --UUAGCCGUUAAGACGACUGCCAAAACAUUCAAUAAAUAUGUUGGCUCAUAUAAAUUAUGGCGGCUAAUGAACAG- --((((((((..........((((..((((.........)))))))).((((.....)))))))))))).......- ( -18.80, z-score = -3.13, R) >droAna3.scaffold_12911 5067740 75 - 5364042 --CUAGCCGUUAAAGCGACUGCCAAAACAUUCAAUAAAUAUGUUGGCUCAUAUAAAUUAUGGCGGCUAAUGAACACC --.(((((((..........((((..((((.........)))))))).((((.....)))))))))))......... ( -18.20, z-score = -2.45, R) >dp4.chr2 2241511 77 - 30794189 CUCGAGCCGUUAAAAUGACUGCCAAAACAUUCAAUAAAUAUGUUGGCUCAUAUAAAUUAUGGCGGCUAAUGAACCCG ....((((((((.(((....((((..((((.........))))))))........))).)))))))).......... ( -17.30, z-score = -2.37, R) >droPer1.super_7 2406338 77 - 4445127 CUCGAGCCGUUAAAAUGACUGCCAAAACAUUCAAUAAAUAUGUUGGCUCAUAUAAAUUAUGGCGGCUAAUGAACCCG ....((((((((.(((....((((..((((.........))))))))........))).)))))))).......... ( -17.30, z-score = -2.37, R) >droWil1.scaffold_181089 5624781 75 - 12369635 --CUAGCCGUUAAAACGACUGCCAAAACAUUCAAUAAAUAUGUUGGCUCAUAUAAAUUAUGGCAGCUAAUGAACACG --...(((((..........((((..((((.........))))))))...........))))).............. ( -13.10, z-score = -1.47, R) >droVir3.scaffold_12855 705520 75 - 10161210 --CGAGCCGUUAAAACGCCUGCCAAAACAUUCAAUAAAUAUAUUGGCUCAUAUAAAUUAUGGCGGCUAAUGAACACG --..((((((..........(((((.................))))).((((.....)))))))))).......... ( -15.23, z-score = -1.68, R) >droMoj3.scaffold_6540 21884923 75 + 34148556 --UGAGCCGUUAAAACGGCUGCCAAAACAUUCAAUAAAUAUGUUGGCUCAUAUAAAUUAUGGCGGCUAAUGAACACG --..((((((....))))))(((........(((((....)))))((.((((.....)))))))))........... ( -20.10, z-score = -2.97, R) >droGri2.scaffold_14906 1809573 75 - 14172833 --CUAGCCGUUAAAGCGACUGCCAAAACAUUCAAUAAAUAUGUUGGUUCAUAUAAAUUAUGGCGGCUAAUGAACACG --.(((((((..........((((..((((.........)))))))).((((.....)))))))))))......... ( -15.90, z-score = -1.67, R) >consensus __CUAGCCGUUAAAACGACUGCCAAAACAUUCAAUAAAUAUGUUGGCUCAUAUAAAUUAUGGCGGCUAAUGAACACG ....((((((..........((((..((((.........)))))))).((((.....)))))))))).......... (-15.53 = -15.54 + 0.02)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:54:59 2011