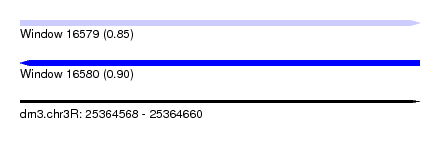
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 25,364,568 – 25,364,660 |
| Length | 92 |
| Max. P | 0.901208 |

| Location | 25,364,568 – 25,364,660 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 67.37 |
| Shannon entropy | 0.55075 |
| G+C content | 0.64257 |
| Mean single sequence MFE | -39.41 |
| Consensus MFE | -21.24 |
| Energy contribution | -22.80 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.848097 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
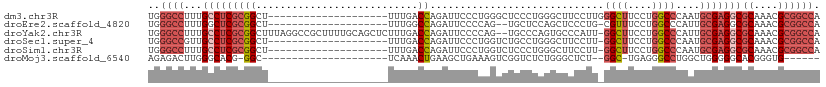
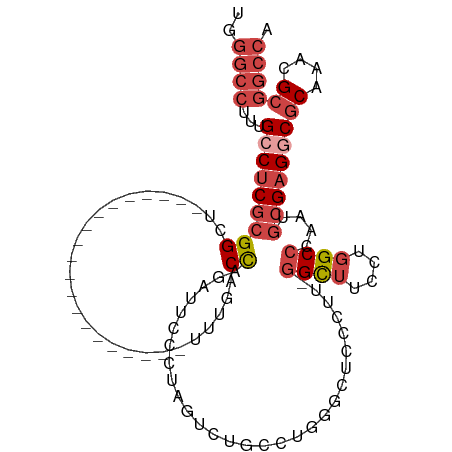
>dm3.chr3R 25364568 92 + 27905053 UGGCCGCGUUUGCGCCUCGCAUUGGGCCAGGAAGCCCAAGGAAGCCCAGGGAGCCCAGGGAAUCUGGUCAAA--------------------AGCCGCGAGGCAAAGGCCCA .((((((....))(((((((.((((((......))))))(((..(((.((....)).)))..)))(((....--------------------.))))))))))...)))).. ( -46.70, z-score = -2.42, R) >droEre2.scaffold_4820 7829678 89 - 10470090 UGGCCGCGUUUGCGCCUCGCAAUGGGCCAGGAAACG-CAGGGAGCUGGAGCA--CUGGGGAAUCUGGCCAAA--------------------AGCCGCGAGCCAAAGGCCCA .(((((((....)))(((((....(((((((...(.-(((...((....)).--))).)...)))))))...--------------------....))))).....)))).. ( -37.50, z-score = -0.11, R) >droYak2.chr3R 7716329 109 - 28832112 UGGCCGCGUUUGCGCCUCGCAAUGGGCCAGGAAGCC-AAUGGGCACUGGGCA--CUGGGGAAUCUGGUCAAAGAGCUGCAAAAGCGGCCUAAAGCCGCGAGGCAAAGGCCCA .((((((....))(((((((....(.((((...(((-....))).)))).).--...........(((...((.(((((....)))))))...))))))))))...)))).. ( -49.50, z-score = -1.11, R) >droSec1.super_4 4222928 91 + 6179234 UGGCCGCGUUUGCGCCUCGCAUUGGGCCAGGAAGCC-AAGGAAGCCCAGGCAGACCAGGGAAUCUGGUCAAA--------------------AGCCGCGAGGCAACGGCCCA .((((((....))(((((((..(((((.........-......)))))(((.((((((.....))))))...--------------------.))))))))))...)))).. ( -45.16, z-score = -2.70, R) >droSim1.chr3R 25037422 91 + 27517382 UGGCCGCGUUUGCGCCUCGCAUUGGGCCAGGAAGCC-AAGGAAGCCCAGGGAGACCAGGGAAUCUGGUCAAA--------------------AGCCGCGAGGCAAAGGCCCA .((((((....))(((((((....(((......)))-..(((..(((.((....)).)))..)))(((....--------------------.))))))))))...)))).. ( -44.00, z-score = -2.48, R) >droMoj3.scaffold_6540 21872705 81 - 34148556 ------CACCCGUGCCCCCAGCCAGGCCCUCA-GCC--AGAGCCCAGAGACCGACUUUCAGCUUCAGUUUGA---------------------GCC-CGUGCCCAAGUCUCU ------..............(((.(((.....-)))--.).))..((((((.........((((......))---------------------)).-.........)))))) ( -13.61, z-score = 1.14, R) >consensus UGGCCGCGUUUGCGCCUCGCAUUGGGCCAGGAAGCC_AAGGAAGCCCAGGCAGACCAGGGAAUCUGGUCAAA____________________AGCCGCGAGGCAAAGGCCCA .((((((....))(((((((....(((......)))...(((..(((.((....)).)))..)))...............................)))))))...)))).. (-21.24 = -22.80 + 1.56)

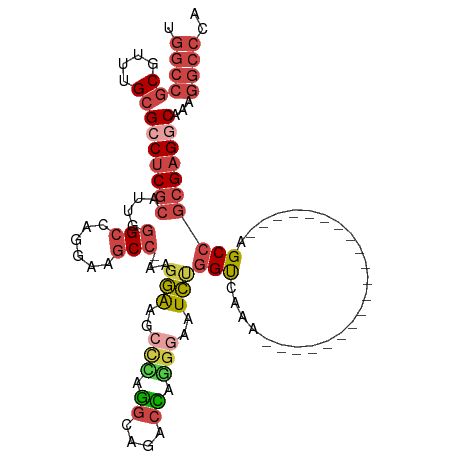
| Location | 25,364,568 – 25,364,660 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 67.37 |
| Shannon entropy | 0.55075 |
| G+C content | 0.64257 |
| Mean single sequence MFE | -41.65 |
| Consensus MFE | -18.70 |
| Energy contribution | -20.48 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.901208 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3R 25364568 92 - 27905053 UGGGCCUUUGCCUCGCGGCU--------------------UUUGACCAGAUUCCCUGGGCUCCCUGGGCUUCCUUGGGCUUCCUGGCCCAAUGCGAGGCGCAAACGCGGCCA ..((((...((((((((((.--------------------...).)).((..(((.((....)).)))..)).(((((((....))))))).)))))))((....)))))). ( -46.50, z-score = -2.49, R) >droEre2.scaffold_4820 7829678 89 + 10470090 UGGGCCUUUGGCUCGCGGCU--------------------UUUGGCCAGAUUCCCCAG--UGCUCCAGCUCCCUG-CGUUUCCUGGCCCAUUGCGAGGCGCAAACGCGGCCA ..((((...(.(((((((..--------------------...((((((......(((--.((....))...)))-......))))))..))))))).)((....)))))). ( -34.70, z-score = 0.14, R) >droYak2.chr3R 7716329 109 + 28832112 UGGGCCUUUGCCUCGCGGCUUUAGGCCGCUUUUGCAGCUCUUUGACCAGAUUCCCCAG--UGCCCAGUGCCCAUU-GGCUUCCUGGCCCAUUGCGAGGCGCAAACGCGGCCA ..((((...((((((((((...((((.((....)).)).))..).)).........((--((.((((.(((....-)))...))))..)))))))))))((....)))))). ( -42.90, z-score = -0.61, R) >droSec1.super_4 4222928 91 - 6179234 UGGGCCGUUGCCUCGCGGCU--------------------UUUGACCAGAUUCCCUGGUCUGCCUGGGCUUCCUU-GGCUUCCUGGCCCAAUGCGAGGCGCAAACGCGGCCA ..(((((((((((((((((.--------------------...((((((.....)))))).)))((((((.....-((...)).))))))..)))))))).....)))))). ( -50.30, z-score = -4.34, R) >droSim1.chr3R 25037422 91 - 27517382 UGGGCCUUUGCCUCGCGGCU--------------------UUUGACCAGAUUCCCUGGUCUCCCUGGGCUUCCUU-GGCUUCCUGGCCCAAUGCGAGGCGCAAACGCGGCCA ..((((...(((((((((..--------------------...((((((.....))))))..))((((((.....-((...)).))))))..)))))))((....)))))). ( -44.40, z-score = -3.14, R) >droMoj3.scaffold_6540 21872705 81 + 34148556 AGAGACUUGGGCACG-GGC---------------------UCAAACUGAAGCUGAAAGUCGGUCUCUGGGCUCU--GGC-UGAGGGCCUGGCUGGGGGCACGGGUG------ .....((..(.((.(-(((---------------------(((....((..(((.....)))..))))))))))--).)-..)).(((((((.....)).))))).------ ( -31.10, z-score = -0.40, R) >consensus UGGGCCUUUGCCUCGCGGCU____________________UUUGACCAGAUUCCCUAGUCUGCCUGGGCUCCCUU_GGCUUCCUGGCCCAAUGCGAGGCGCAAACGCGGCCA ..((((...(((((((((...........................)).......((((...(((.(((....))).)))...))))......)))))))((....)))))). (-18.70 = -20.48 + 1.78)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:54:56 2011