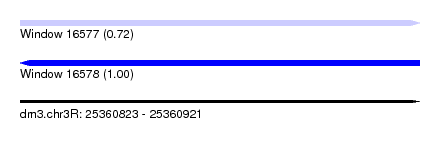
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 25,360,823 – 25,360,921 |
| Length | 98 |
| Max. P | 0.995556 |

| Location | 25,360,823 – 25,360,921 |
|---|---|
| Length | 98 |
| Sequences | 11 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 80.45 |
| Shannon entropy | 0.36168 |
| G+C content | 0.46353 |
| Mean single sequence MFE | -31.14 |
| Consensus MFE | -17.68 |
| Energy contribution | -16.67 |
| Covariance contribution | -1.01 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.723570 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 25360823 98 + 27905053 -------------GGUCGCGGAACGUGGCCGCU-----UGUUGGCUUGUUGUCUUUGUUAGUGGUAGCAGUUAGUUGACUGCACUUACCACUAAGUUAAGAUGUUUUUGUCUCCGC--- -------------....((((((((.(((((..-----...)))))....(((((..(((((((((((((((....))))))...)))))))))...))))).....))).)))))--- ( -34.70, z-score = -2.92, R) >droAna3.scaffold_12911 5055121 101 + 5364042 -------------GGUCGCGGAACGUGGCCGUUGGC--UGUUGGCUUGUUGUCUUUGUUAGUGGUAGCAGUUAGUUGACUGCACUUACCACUAAGUUAAGAUGUUUUUGUCUCCGC--- -------------....(((((..(..(((...)))--..).(((....((((((..(((((((((((((((....))))))...)))))))))...)))))).....))))))))--- ( -35.80, z-score = -2.81, R) >droEre2.scaffold_4820 7825855 98 - 10470090 -------------GGUCGCGGAACGUGGCCGCU-----UGUUGGCUUGUUGUCUUUGUUAGUGGUAGCAGUUAGUUGACUGCACUUACCACUAAGUUAAGAUGUUUUUGUCUCCGC--- -------------....((((((((.(((((..-----...)))))....(((((..(((((((((((((((....))))))...)))))))))...))))).....))).)))))--- ( -34.70, z-score = -2.92, R) >droYak2.chr3R 7712421 98 - 28832112 -------------GGUCGCGGAACGUGGCCGCU-----UGUUGGCUUGUUGUCUUUGUUAGUGGUAGCAGUUAGUUGACUGCACUUACCACUAAGUUAAGAUGUUUUUGUCUCCGC--- -------------....((((((((.(((((..-----...)))))....(((((..(((((((((((((((....))))))...)))))))))...))))).....))).)))))--- ( -34.70, z-score = -2.92, R) >droSec1.super_4 4219156 90 + 6179234 -------------GGUCGCGGAACGUGGCCGCU-------------UGUUGUCUUUGUUAGUGGUAGCAGUUAGUUGACUGCACUUACCACUAAGUUAAGAUGUUUUUGUCUCCGC--- -------------(((((((...)))))))...-------------....(((((..(((((((((((((((....))))))...)))))))))...)))))..............--- ( -31.50, z-score = -2.91, R) >droSim1.chr3R 25033704 98 + 27517382 -------------GGUCGCGGAACGUGGCCGCU-----UGUUGGCUUGUUGUCUUUGUUAGUGGUAGCAGUUAGUUGACUGCACUUACCACUAAGUUAAGAUGUUUUUGUCUCCGC--- -------------....((((((((.(((((..-----...)))))....(((((..(((((((((((((((....))))))...)))))))))...))))).....))).)))))--- ( -34.70, z-score = -2.92, R) >droGri2.scaffold_14906 1794101 101 + 14172833 ---------------GCUUGAGGCCUGGCCUGGCACUCUUUUCGUUUGUUGUCUUUGUUAGUGGUAGCAGUUAGUUGACUACACUUACCACUAAGUUAAGAUGUUUU-GUCUUCGGU-- ---------------....((((((......))).)))............(((((..(((((((((..((((((....))).))))))))))))...))))).....-.........-- ( -25.40, z-score = -0.37, R) >droMoj3.scaffold_6540 21867522 95 - 34148556 ---------------GCUUGAGGCCUGGCACUG------UUUCGCUUGUUGUCUUUGUUAGUGGUAGCAGUUAGUUGGUUGCACUUACCACUAAGUUAAGAUGUUUU-GUCUUGGGC-- ---------------(((..((((..(((....------....)))....(((((..(((((((((((((........))))...)))))))))...))))).....-))))..)))-- ( -28.70, z-score = -1.32, R) >droVir3.scaffold_12855 689416 95 + 10161210 ---------------GCUUGAGGCCUGGCACUG------UUUUGUUUGUUGUCUUUGUUAGUGGUAGCAGUUAGUUGGCUGCACUUACCACUAAGUUAAGAUGUUUU-GUCUUGGGC-- ---------------(((..((((..(((((..------....)).....(((((..(((((((((((((((....))))))...)))))))))...))))))))..-))))..)))-- ( -33.50, z-score = -2.91, R) >droPer1.super_7 2392397 105 + 4445127 UGGAUGUGGAUGCGGACCUGGAACGUGGCCGGU-------------UGUUGUCUUUGUUAGUGGUAGCAGUUAGUUGACUUCACUUACCACUAAGUCA-GAUGUUUUUGUCUUUGCUGU .(((((..(((.(((.((........)))))..-------------....((((...((((((((((.((((....))))....))))))))))...)-))))))..)))))....... ( -24.40, z-score = 0.93, R) >dp4.chr2 2225220 100 + 30794189 ------UGGAUGUGGACCUGGAACGUGGCCGGU-------------UGUUGUCUUUGUUAGUGGUAGCAGUUAGUUGACUUCACUUACCACUAAGUUAAGAUGUUUUUGUCUUUGCUGU ------.(((((..((((.((.......)))))-------------)..)))))....(((((((((.((((....))))....)))))))))(((.(((((......))))).))).. ( -24.40, z-score = -0.17, R) >consensus _____________GGUCGCGGAACGUGGCCGCU_____UGUUGGCUUGUUGUCUUUGUUAGUGGUAGCAGUUAGUUGACUGCACUUACCACUAAGUUAAGAUGUUUUUGUCUCCGC___ .................(((((((..(((..................)))(((((..(((((((((..((((((....))).))))))))))))...)))))......)).)))))... (-17.68 = -16.67 + -1.01)


| Location | 25,360,823 – 25,360,921 |
|---|---|
| Length | 98 |
| Sequences | 11 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 80.45 |
| Shannon entropy | 0.36168 |
| G+C content | 0.46353 |
| Mean single sequence MFE | -26.15 |
| Consensus MFE | -15.84 |
| Energy contribution | -16.32 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.12 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.82 |
| SVM RNA-class probability | 0.995556 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 25360823 98 - 27905053 ---GCGGAGACAAAAACAUCUUAACUUAGUGGUAAGUGCAGUCAACUAACUGCUACCACUAACAAAGACAACAAGCCAACA-----AGCGGCCACGUUCCGCGACC------------- ---((((((.(.......((((...(((((((((...(((((......))))))))))))))..))))......(((....-----...)))...)))))))....------------- ( -31.50, z-score = -4.65, R) >droAna3.scaffold_12911 5055121 101 - 5364042 ---GCGGAGACAAAAACAUCUUAACUUAGUGGUAAGUGCAGUCAACUAACUGCUACCACUAACAAAGACAACAAGCCAACA--GCCAACGGCCACGUUCCGCGACC------------- ---((((((.(.......((((...(((((((((...(((((......))))))))))))))..))))......(((....--......)))...)))))))....------------- ( -30.90, z-score = -4.43, R) >droEre2.scaffold_4820 7825855 98 + 10470090 ---GCGGAGACAAAAACAUCUUAACUUAGUGGUAAGUGCAGUCAACUAACUGCUACCACUAACAAAGACAACAAGCCAACA-----AGCGGCCACGUUCCGCGACC------------- ---((((((.(.......((((...(((((((((...(((((......))))))))))))))..))))......(((....-----...)))...)))))))....------------- ( -31.50, z-score = -4.65, R) >droYak2.chr3R 7712421 98 + 28832112 ---GCGGAGACAAAAACAUCUUAACUUAGUGGUAAGUGCAGUCAACUAACUGCUACCACUAACAAAGACAACAAGCCAACA-----AGCGGCCACGUUCCGCGACC------------- ---((((((.(.......((((...(((((((((...(((((......))))))))))))))..))))......(((....-----...)))...)))))))....------------- ( -31.50, z-score = -4.65, R) >droSec1.super_4 4219156 90 - 6179234 ---GCGGAGACAAAAACAUCUUAACUUAGUGGUAAGUGCAGUCAACUAACUGCUACCACUAACAAAGACAACA-------------AGCGGCCACGUUCCGCGACC------------- ---(((((..........((((...(((((((((...(((((......))))))))))))))..)))).....-------------.(((....))))))))....------------- ( -29.00, z-score = -4.26, R) >droSim1.chr3R 25033704 98 - 27517382 ---GCGGAGACAAAAACAUCUUAACUUAGUGGUAAGUGCAGUCAACUAACUGCUACCACUAACAAAGACAACAAGCCAACA-----AGCGGCCACGUUCCGCGACC------------- ---((((((.(.......((((...(((((((((...(((((......))))))))))))))..))))......(((....-----...)))...)))))))....------------- ( -31.50, z-score = -4.65, R) >droGri2.scaffold_14906 1794101 101 - 14172833 --ACCGAAGAC-AAAACAUCUUAACUUAGUGGUAAGUGUAGUCAACUAACUGCUACCACUAACAAAGACAACAAACGAAAAGAGUGCCAGGCCAGGCCUCAAGC--------------- --.........-......((((...(((((((((...(((((......))))))))))))))..)))).............(((.(((......))))))....--------------- ( -25.00, z-score = -2.37, R) >droMoj3.scaffold_6540 21867522 95 + 34148556 --GCCCAAGAC-AAAACAUCUUAACUUAGUGGUAAGUGCAACCAACUAACUGCUACCACUAACAAAGACAACAAGCGAAA------CAGUGCCAGGCCUCAAGC--------------- --(((......-......((((...(((((((((...(((..........))))))))))))..))))......(((...------...)))..))).......--------------- ( -17.80, z-score = -0.78, R) >droVir3.scaffold_12855 689416 95 - 10161210 --GCCCAAGAC-AAAACAUCUUAACUUAGUGGUAAGUGCAGCCAACUAACUGCUACCACUAACAAAGACAACAAACAAAA------CAGUGCCAGGCCUCAAGC--------------- --(((......-......((((...(((((((((...((((........)))))))))))))..))))......((....------..))....))).......--------------- ( -19.50, z-score = -1.49, R) >droPer1.super_7 2392397 105 - 4445127 ACAGCAAAGACAAAAACAUC-UGACUUAGUGGUAAGUGAAGUCAACUAACUGCUACCACUAACAAAGACAACA-------------ACCGGCCACGUUCCAGGUCCGCAUCCACAUCCA ...((.............((-(...((((((((((((..((....)).)))..)))))))))...))).....-------------...((((........)))).))........... ( -21.30, z-score = -1.34, R) >dp4.chr2 2225220 100 - 30794189 ACAGCAAAGACAAAAACAUCUUAACUUAGUGGUAAGUGAAGUCAACUAACUGCUACCACUAACAAAGACAACA-------------ACCGGCCACGUUCCAGGUCCACAUCCA------ ..................((((...((((((((((((..((....)).)))..)))))))))..)))).....-------------...((((........))))........------ ( -18.10, z-score = -1.07, R) >consensus ___GCGGAGACAAAAACAUCUUAACUUAGUGGUAAGUGCAGUCAACUAACUGCUACCACUAACAAAGACAACAAGCCAACA_____AGCGGCCACGUUCCGCGACC_____________ ..................((((...(((((((((...(((((......))))))))))))))..))))................................................... (-15.84 = -16.32 + 0.48)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:54:55 2011