| Sequence ID | dm3.chr2L |
|---|---|
| Location | 9,114,444 – 9,114,534 |
| Length | 90 |
| Max. P | 0.959602 |

| Location | 9,114,444 – 9,114,534 |
|---|---|
| Length | 90 |
| Sequences | 10 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 69.27 |
| Shannon entropy | 0.62663 |
| G+C content | 0.49932 |
| Mean single sequence MFE | -24.00 |
| Consensus MFE | -12.16 |
| Energy contribution | -11.92 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.959602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 9114444 90 - 23011544 -GGGGGAAUC--------UUGGCUGAUCUUAUCCUCCCCAGACCAGCCAGACAAUAACUAUAAUUGUCGCAUCAGCAUUUUUUC-CGGCCUCCUCCUCAG -((((((...--------.((((((.(((..........))).))))))((((((.......))))))((....))........-.....)))))).... ( -26.20, z-score = -2.61, R) >droSim1.chr2L 8894596 90 - 22036055 -GGGGGAAUC--------UUGGCUGAUCUUAUCCACCCCAGACCAGCCAGACAAUAACUAUAAUUGUCGCAUCAGCAUUUUUUC-CGGCCUCCGCCUCAG -((..(((..--------.((((((.(((..........))).))))))((((((.......))))))((....))..)))..)-)(((....))).... ( -26.50, z-score = -2.63, R) >droSec1.super_3 4580656 87 - 7220098 -GGGGGAAUC--------UUGGCUGAUCUUAUCCACCCCAGACCAGCCAGACAAUAACUAUAAUUGUCGCAUCAGCAUUUUUUC-CGGCCUCCGCCU--- -((..(((..--------.((((((.(((..........))).))))))((((((.......))))))((....))..)))..)-)(((....))).--- ( -26.50, z-score = -2.48, R) >droEre2.scaffold_4929 9718990 85 - 26641161 -GGGGGAAUC--------UUGGCUGAUCUUAUGCGCCCCAGACCAGCCAGACAAUAACUAUAAUUGUCGCAUCAGCA-UUUUUC-CGGCCUCCGCC---- -((..((((.--------.((((((.(((..........))).))))))((((((.......))))))((....)))-)))..)-)(((....)))---- ( -26.00, z-score = -1.60, R) >droYak2.chr2L 11780703 90 - 22324452 -GGGGGAAUC--------UUGGCUGAUCUUAUCCUACCCAGACCAGCCAGACAAUAACUAUAAUUGUCGCAUCAGCAUUUUUUC-CGGCCUCCGCCUCAA -((..(((..--------.((((((.(((..........))).))))))((((((.......))))))((....))..)))..)-)(((....))).... ( -26.50, z-score = -2.62, R) >droAna3.scaffold_12943 462008 98 - 5039921 -GGGGGAGUCGAGACCCUCCGAGUUGUGUUCCCCUGCCCAGACCAGCCAGACAAUAACUAUAAUUGUCGCAUCAGCAUUUUUUC-CGGUCUUAGCUCCAG -(((((((.((.(((.......))).)))))))))((..(((((.....((((((.......))))))((....))........-.)))))..))..... ( -28.80, z-score = -2.03, R) >droWil1.scaffold_180708 12520771 82 + 12563649 ------AGUCUACUCCCACUGGCUCAUCUU------CCUGGUUAGGCCAGACAAUAACUAUAAUUGUCGCAUCAGCGUUUUUUCUCAGCCUCAA------ ------..............((((......------..(((.....)))((((((.......))))))((....))..........))))....------ ( -13.80, z-score = -0.62, R) >droVir3.scaffold_12963 461046 84 - 20206255 ---GGAGAUGGG------AUGGCUGGAGAUUUCCCCGGUUGGUUAGGCAGACAAUAACUAUAAUUGUCGCAUCAGCGUUUU-UG-CUGCUGCUGU----- ---.........------..((((((.(....).))))))......(((((((((.......))))))(((.(((((....-))-))).))))))----- ( -22.30, z-score = 0.22, R) >droMoj3.scaffold_6500 11960076 81 - 32352404 ------------------GGAGUAGGAAAUCUCCCCGCUUGGUUAGGCAGACAAUAACUAUAAUUGUCGCAUCAGCGUUUUGUG-CUGCCGCUGCUUUCG ------------------.(((..((.......))..)))....(((((((((((.......))))))((..(((((.....))-)))..)))))))... ( -21.00, z-score = -0.29, R) >droGri2.scaffold_15126 5243365 87 + 8399593 GGAGAGAGUUGG------AUGGCAGAAGACUUCCCCGCUUGGUUAGGCAGACAAUAACUAUAAUUGUCGCAUCAGCGUUUU-UG-CUGCUGUCGG----- ...........(------(((((((.((((......(((......))).((((((.......))))))((....)))))).-..-))))))))..----- ( -22.40, z-score = 0.24, R) >consensus _GGGGGAAUC________UUGGCUGAUCUUAUCCCCCCCAGACCAGCCAGACAAUAACUAUAAUUGUCGCAUCAGCAUUUUUUC_CGGCCUCCGCCU___ ....................(((((........................((((((.......))))))((....)).........))))).......... (-12.16 = -11.92 + -0.24)

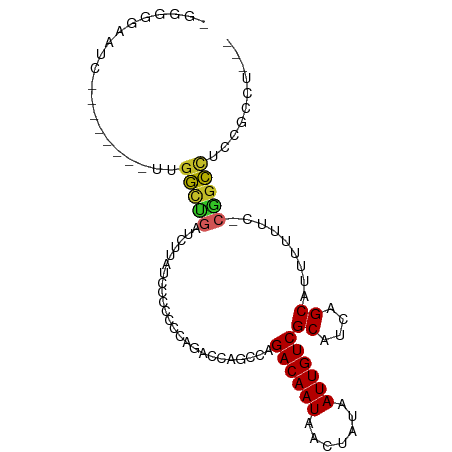

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:27:33 2011