| Sequence ID | dm3.chr3R |
|---|---|
| Location | 25,352,558 – 25,352,650 |
| Length | 92 |
| Max. P | 0.934081 |

| Location | 25,352,558 – 25,352,650 |
|---|---|
| Length | 92 |
| Sequences | 11 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 81.33 |
| Shannon entropy | 0.35540 |
| G+C content | 0.41078 |
| Mean single sequence MFE | -27.06 |
| Consensus MFE | -16.93 |
| Energy contribution | -16.74 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.934081 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
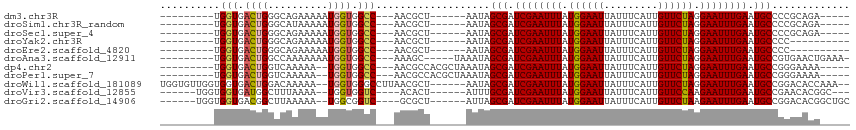
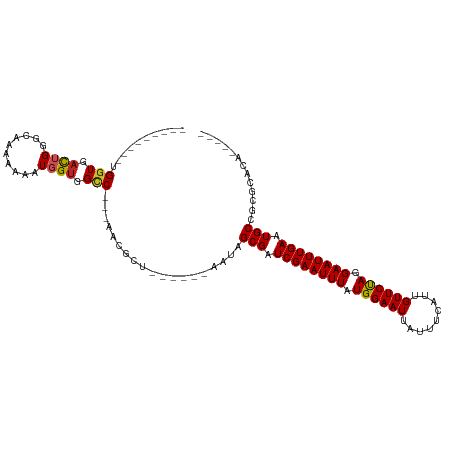
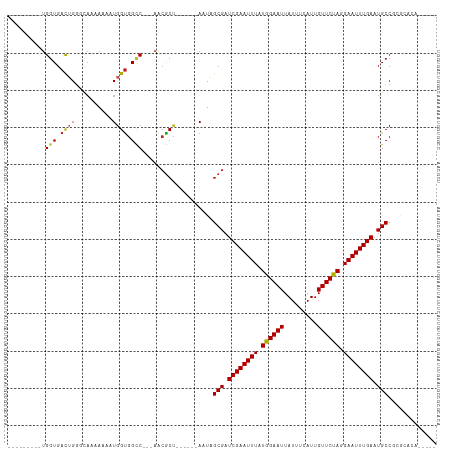
>dm3.chr3R 25352558 92 + 27905053 ---------UGGUGACUGGGCAGAAAAAUGGUGGCC---AACGCU------AAUAGCGAUCGAAUUUAUGGAAUUAUUUCAUUGUUCUAGGAAUUUGAAUGCCCCGCAGA----- ---------..(((...(((((......(((((...---..))))------).......((((((((.((((((.........)))))).)))))))).))))))))...----- ( -25.30, z-score = -1.42, R) >droSim1.chr3R_random 1236501 92 + 1307089 ---------UGGUGACUGGGCAUAAAAAUGGUGGCC---AACGCU------AAUAGCGAUCGAAUUUAUGGAAUUAUUUCAUUGUUCUAGGAAUUUGAAUGCCCCGCAGA----- ---------..(((...((((((.....(((((...---..))))------).......((((((((.((((((.........)))))).)))))))))))))))))...----- ( -26.30, z-score = -1.82, R) >droSec1.super_4 4211046 92 + 6179234 ---------UGGUGACUGGGCAGAAAAAUGGUGGCC---AACGCU------AAUAGCGAUCGAAUUUAUGGAAUUAUUUCAUUGUUCUAGGAAUUUGAAUGCCCCGCAGA----- ---------..(((...(((((......(((((...---..))))------).......((((((((.((((((.........)))))).)))))))).))))))))...----- ( -25.30, z-score = -1.42, R) >droYak2.chr3R 7704233 87 - 28832112 ---------UGGUGACUGGGCAGAAAAAUGGUGGCC---AACGCU------AAUAGCGAUCGAAUUUAUGGAAUUAUUUCAUUGUUCUAGGAAUUUGAAUGCCCC---------- ---------.(....).(((((......(((((...---..))))------).......((((((((.((((((.........)))))).)))))))).))))).---------- ( -24.70, z-score = -2.09, R) >droEre2.scaffold_4820 7817790 87 - 10470090 ---------UGGUGACUGGGCAGAAAAAUGGUGGCC---AACGCU------AAUAGCGAUCGAAUUUAUGGAAUUAUUUCAUUGUUCUAGGAAUUUGAAUGCCCC---------- ---------.(....).(((((......(((((...---..))))------).......((((((((.((((((.........)))))).)))))))).))))).---------- ( -24.70, z-score = -2.09, R) >droAna3.scaffold_12911 5046965 97 + 5364042 ---------UGGUGACUGGCCAAAAAAAUGGUGGCC---AAAGC-----UAAAUAGCGAUCGAAUUUAUGGAAUUAUUUCAUUGUUCUAGGAAUUUGAAUGCCGUGAACUGAAA- ---------((((...((((((.........)))))---)..))-----))....(((.((((((((.((((((.........)))))).)))))))).)))............- ( -25.30, z-score = -2.54, R) >dp4.chr2 2216786 96 + 30794189 ---------UGGUGACUGGUCAAAAA--UGGUGGCC---AACGCCACGCUAAAUAGCGAUCGAAUUUAUGGAAUUAUUUCAUUGUUCUAGGAAUUUGAAUGCCGGGAAAA----- ---------......(((((......--(((((...---..)))))((((....)))).((((((((.((((((.........)))))).))))))))..))))).....----- ( -27.10, z-score = -2.06, R) >droPer1.super_7 2383979 96 + 4445127 ---------UGGUGACUGGUCAAAAA--UGGUGGCC---AACGCCACGCUAAAUAGCGAUCGAAUUUAUGGAAUUAUUUCAUUGUUCUAGGAAUUUGAAUGCCGGGAAAA----- ---------......(((((......--(((((...---..)))))((((....)))).((((((((.((((((.........)))))).))))))))..))))).....----- ( -27.10, z-score = -2.06, R) >droWil1.scaffold_181089 5597672 105 + 12369635 UGGUGUUGGUGGUGACUGGACAAAAA--UGGUGGGCCUUAACGCU------AAUAGCGAUCGAAUUUAUGGAAUUAUUUCAUUGUUCUAGGAAUUUGAAUGCCGGACACCAAA-- (((((((.(.(....)..........--(((((........))))------)...(((.((((((((.((((((.........)))))).)))))))).)))).)))))))..-- ( -29.70, z-score = -2.13, R) >droVir3.scaffold_12855 681023 94 + 10161210 ------UGGUGGUGAUGGCUUUAAAA--UGGUGGUC----ACACU------AUUUGCGAUCGAAUUUAUGGAAUUAUUUCAUUGUUCCAAGAAUUUGAAUGCCGAACACGGC--- ------..((.(((.((((....(((--(((((...----.))))------))))....((((((((.((((((.........)))))).))))))))..))))..))).))--- ( -29.60, z-score = -3.28, R) >droGri2.scaffold_14906 1785951 97 + 14172833 ------UGGUGGUGACGGCUUAAAAA--UGGCGGUC----GCGCU------AUUAGCGAUCGAAUUUAUGGAAUUAUUUCAUUGUUCUAAGAAUUUGAAUGCCGGACACGGCUGC ------.(((.(((.((((.....((--(((((...----.))))------))).....((((((((.((((((.........)))))).))))))))..))))..))).))).. ( -32.60, z-score = -3.31, R) >consensus _________UGGUGACUGGGCAAAAAAAUGGUGGCC___AACGCU______AAUAGCGAUCGAAUUUAUGGAAUUAUUUCAUUGUUCUAGGAAUUUGAAUGCCGCGCACA_____ .............................((((........))))..........(((.((((((((.((((((.........)))))).)))))))).)))............. (-16.93 = -16.74 + -0.20)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:54:53 2011