
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 25,339,547 – 25,339,668 |
| Length | 121 |
| Max. P | 0.995665 |

| Location | 25,339,547 – 25,339,668 |
|---|---|
| Length | 121 |
| Sequences | 4 |
| Columns | 133 |
| Reading direction | forward |
| Mean pairwise identity | 81.28 |
| Shannon entropy | 0.29060 |
| G+C content | 0.55718 |
| Mean single sequence MFE | -49.75 |
| Consensus MFE | -39.71 |
| Energy contribution | -39.53 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.983400 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
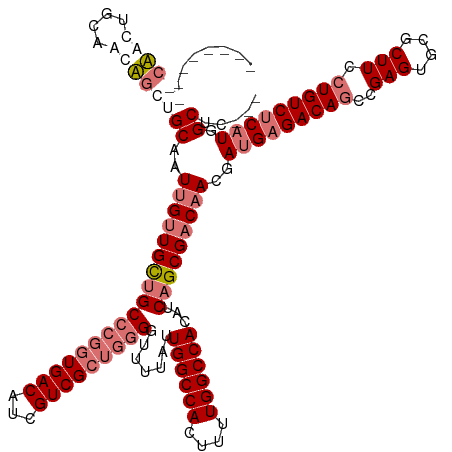
>dm3.chr3R 25339547 121 + 27905053 ---GAGCCAUGAGACAGGAAGCGCACUCGGCUGUCUCAUCGUUGUCGCUGAUGUGGCCAAAAGUGGCCAAUAAAACCCCAGCGACGAUGUCACCGGGCAGCAACAAUUGCAGUUGCUGUUGCAG--------- ---..((((((((((((.............)))))))))(((((((((((...((((((....)))))).........)))))))))))......))).((((((...((....))))))))..--------- ( -49.62, z-score = -2.98, R) >droSim1.chr3R 25012867 130 + 27517382 ---GAGCCAUGAGACAGGAAGCGCACUCGGCUGUCUCAUCGUUGUCGCUGAUGUGGCCAAAAGUGGCCAAUAAAACCCCAGCGACGAUGUCACCGGGCAGCAACAAUUGCAGCAGUUGCAGUUGCUGUUGCAG ---...(((((((((((.............)))))))))(((((((((((...((((((....)))))).........))))))))))).....))((((((.(((((((((...))))))))).)))))).. ( -55.92, z-score = -3.46, R) >droSec1.super_4 4198200 130 + 6179234 ---GAGCCAUGAGACAGGAAGCGCACUCGGCUGUCUCAUCUUUGUCGCUGAUGUGGCCAAAAGUGGCCAAUAAAACCCCAGCGACGAUGUCACCGGGCAGCAACAAUUGCAGCAGUUGCAGUUGCUCUUGCAG ---..((((((((((((.............)))))))))..(((((((((...((((((....)))))).........)))))))))........))).(((((((((((((...)))))))))...)))).. ( -49.52, z-score = -2.01, R) >dp4.chr2 2202792 120 + 30794189 GUUCGGCUAGGGGACAGGAAGCUGGCUCGCUUGUCUCAUCGUUGUCGCUGAUGUGGCCAAAAGUGGCCAAUAAAACCCAAGCGACGAUGUCA----GCCACAGCCACUGCCACUGCCACAGCAG--------- ....((((..((((((((.((....))..))))))))..((((((((((....((((((....))))))..........))))))))))..)----))).............((((....))))--------- ( -43.94, z-score = -1.20, R) >consensus ___GAGCCAUGAGACAGGAAGCGCACUCGGCUGUCUCAUCGUUGUCGCUGAUGUGGCCAAAAGUGGCCAAUAAAACCCCAGCGACGAUGUCACCGGGCAGCAACAAUUGCAGCAGCUGCAGCAG_________ .....((((((((((((..((....))...)))))))))(((((((((((...((((((....)))))).........)))))))))))......))).((((...)))).(((((....)))))........ (-39.71 = -39.53 + -0.19)



| Location | 25,339,547 – 25,339,668 |
|---|---|
| Length | 121 |
| Sequences | 4 |
| Columns | 133 |
| Reading direction | reverse |
| Mean pairwise identity | 81.28 |
| Shannon entropy | 0.29060 |
| G+C content | 0.55718 |
| Mean single sequence MFE | -52.33 |
| Consensus MFE | -39.86 |
| Energy contribution | -43.92 |
| Covariance contribution | 4.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.83 |
| SVM RNA-class probability | 0.995665 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 25339547 121 - 27905053 ---------CUGCAACAGCAACUGCAAUUGUUGCUGCCCGGUGACAUCGUCGCUGGGGUUUUAUUGGCCACUUUUGGCCACAUCAGCGACAACGAUGAGACAGCCGAGUGCGCUUCCUGUCUCAUGGCUC--- ---------.((((........)))).((((((((((((((((((...))))))))).......((((((....))))))...)))))))))(.(((((((((..(((....))).))))))))).)...--- ( -53.10, z-score = -4.07, R) >droSim1.chr3R 25012867 130 - 27517382 CUGCAACAGCAACUGCAACUGCUGCAAUUGUUGCUGCCCGGUGACAUCGUCGCUGGGGUUUUAUUGGCCACUUUUGGCCACAUCAGCGACAACGAUGAGACAGCCGAGUGCGCUUCCUGUCUCAUGGCUC--- .((((.(((.........))).)))).((((((((((((((((((...))))))))).......((((((....))))))...)))))))))(.(((((((((..(((....))).))))))))).)...--- ( -56.00, z-score = -3.85, R) >droSec1.super_4 4198200 130 - 6179234 CUGCAAGAGCAACUGCAACUGCUGCAAUUGUUGCUGCCCGGUGACAUCGUCGCUGGGGUUUUAUUGGCCACUUUUGGCCACAUCAGCGACAAAGAUGAGACAGCCGAGUGCGCUUCCUGUCUCAUGGCUC--- ......((((...((((.....)))).((((((((((((((((((...))))))))).......((((((....))))))...)))))))))..(((((((((..(((....))).))))))))).))))--- ( -56.90, z-score = -3.81, R) >dp4.chr2 2202792 120 - 30794189 ---------CUGCUGUGGCAGUGGCAGUGGCUGUGGC----UGACAUCGUCGCUUGGGUUUUAUUGGCCACUUUUGGCCACAUCAGCGACAACGAUGAGACAAGCGAGCCAGCUUCCUGUCCCCUAGCCGAAC ---------......((((((.(((((.((((((.((----(....((((((.(((.(((....((((((....))))))....)))..)))))))))....))).)..)))))..))).)).)).))))... ( -43.30, z-score = -0.51, R) >consensus _________CAACUGCAACAGCUGCAAUUGUUGCUGCCCGGUGACAUCGUCGCUGGGGUUUUAUUGGCCACUUUUGGCCACAUCAGCGACAACGAUGAGACAGCCGAGUGCGCUUCCUGUCUCAUGGCUC___ .......((((((....))))))((..((((((((((((((((((...))))))))).......((((((....))))))...)))))))))..(((((((((..(((....))).))))))))).))..... (-39.86 = -43.92 + 4.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:54:51 2011