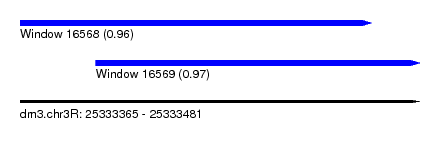
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 25,333,365 – 25,333,481 |
| Length | 116 |
| Max. P | 0.967644 |

| Location | 25,333,365 – 25,333,467 |
|---|---|
| Length | 102 |
| Sequences | 10 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 79.08 |
| Shannon entropy | 0.39615 |
| G+C content | 0.49707 |
| Mean single sequence MFE | -31.49 |
| Consensus MFE | -26.19 |
| Energy contribution | -26.10 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.964939 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 25333365 102 + 27905053 ---------CUGGAGA-GCUGCGCCUGCUCC--AAGUAAUGGGCUGCCGUUAAAGGUGGCGACCGGGAGCCGCCUAUUCCGACAAGUGAUUGUUUUAUAAUUGCAAUCGCAAUU ---------.((((((-((.((((((((...--..))...)))).)).)))..(((((((........))))))).)))))....((((((((.........)))))))).... ( -34.90, z-score = -1.27, R) >droAna3.scaffold_12911 5027749 84 + 5364042 ----------------------------CAG--AAAUAAUGGGCAGCCGUUAAAGGUGGCGACCAGGAGCCGCCUAGCCCGACAAGUGAUUGUUUUAUAAUUGCAAUCGCAAUU ----------------------------...--......(((((.........(((((((........))))))).)))))....((((((((.........)))))))).... ( -27.60, z-score = -2.64, R) >droEre2.scaffold_4820 7798206 112 - 10470090 CUGGAGAUGCUGGAGAUGCUGGGCAGGCCGC--GAGUAAUGGGCUGCCGUUAAAGGUGGCGACCAGGAGCCGCCUAGCCCGACAAGUGAUUGUUUUAUAAUUGCAGUCGCAAUU (..(.....)..)...((.(((((((((.((--......(((((..((......))..))..)))...)).)))).))))).)).((((((((.........)))))))).... ( -41.40, z-score = -1.28, R) >droSec1.super_4 4191955 102 + 6179234 ---------CUGGAGA-GCUGCGCCUGCUGC--AAGUAAUGGGUUGCCGUUAAAGGUGGCGACCGGGAGCCGCCUAUACCGACAAGUGAUUGUUUUAUAAUUGCAAUCGCAAUU ---------.(((...-((.((....)).))--...((((((....)))))).(((((((........)))))))...)))....((((((((.........)))))))).... ( -32.40, z-score = -0.65, R) >droSim1.chr3R 25006711 102 + 27517382 ---------CUGGAGA-GCUGCGCCUGCUGC--AAGUAAUGGGCUGCCGUUAAAGGUGGCGACCGGGAGCCGCCUAUCCCGACAAGUGAUUGUUUUAUAAUUGCAAUCGCAAUU ---------..(((.(-((.((((((..((.--...))..)))).)).)))..(((((((........))))))).)))......((((((((.........)))))))).... ( -34.70, z-score = -0.88, R) >droPer1.super_7 2364055 104 + 4445127 ---------CUGGAAAAAA-GGAAGGAAUACGAAAAUAAUGGGGAGCCGUUAAAGGUGGCGACCGGGAGCCGCCUAGCCCGACAAGUGAUUGUUUUAUAAUUGCAAUCGCAAUU ---------..........-....((..........((((((....)))))).(((((((........)))))))...)).....((((((((.........)))))))).... ( -30.50, z-score = -1.97, R) >dp4.chr2 2196926 105 + 30794189 ---------CUGGAAAAAAAGGAAGGAAUACGAAAAUAAUGGGGAGCCGUUAAAGGUGGCGACCGGGAGCCGCCUAGCCCGACAAGUGAUUGUUUUAUAAUUGCAAUCGCAAUU ---------...............((..........((((((....)))))).(((((((........)))))))...)).....((((((((.........)))))))).... ( -30.50, z-score = -1.97, R) >droVir3.scaffold_12855 657810 82 + 10161210 -------------------------------CAAAAUAAUGGGCAGCCGUUAA-GGUGGCGACCGGGAGCCGCCUAGCCGGACAAGUGAUUGUUUUAUAAUUGCAAUCGCAAUU -------------------------------.....((((((....))))))(-((((((........)))))))..........((((((((.........)))))))).... ( -27.00, z-score = -1.78, R) >droMoj3.scaffold_6540 21834054 82 - 34148556 -------------------------------CAAAAUAAUGGCCAGCCGUUAA-GGUGGCGACCGGGAGCCGCCUAGCCGGACAAGUGAUUGUUUUAUAAUUGCAAUCGCAAUU -------------------------------.((((((((.((..((((...(-((((((........)))))))...))).)..)).))))))))..((((((....)))))) ( -28.50, z-score = -2.39, R) >droGri2.scaffold_14906 1766450 82 + 14172833 -------------------------------CAAAAUAAUGGGCUGCCGUUAA-GGUGGCGACCGGGAGCCGCCUAGCCGGACAAGUGAUUGUUUUAUAAUUGCAAUCGCAAUU -------------------------------.((((((((.(..(((((...(-((((((........)))))))...))).))..).))))))))..((((((....)))))) ( -27.40, z-score = -1.70, R) >consensus _________CUGGA_A____G_______U_C__AAAUAAUGGGCAGCCGUUAAAGGUGGCGACCGGGAGCCGCCUAGCCCGACAAGUGAUUGUUUUAUAAUUGCAAUCGCAAUU ....................................((((((....))))))..((((((........))))))...........((((((((.........)))))))).... (-26.19 = -26.10 + -0.09)

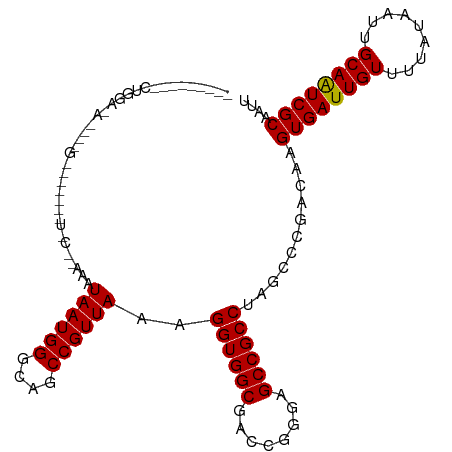
| Location | 25,333,387 – 25,333,481 |
|---|---|
| Length | 94 |
| Sequences | 10 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 83.34 |
| Shannon entropy | 0.31880 |
| G+C content | 0.49065 |
| Mean single sequence MFE | -32.50 |
| Consensus MFE | -26.55 |
| Energy contribution | -26.38 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.967644 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 25333387 94 + 27905053 AGUAAUGGGCUGCCGUUAAAGGUGGCGACCGGGAGCCGCCUAUUCCGACAAGUGAUUGUUUUAUAAUUGCAAUCGCAAUUGCAGCUUGCAGCUU--------------- (((..((((((((((..(.(((((((........))))))).)..))....((((((((.........))))))))....))))))))..))).--------------- ( -35.10, z-score = -2.38, R) >droAna3.scaffold_12911 5027753 80 + 5364042 AAUAAUGGGCAGCCGUUAAAGGUGGCGACCAGGAGCCGCCUAGCCCGACAAGUGAUUGUUUUAUAAUUGCAAUCGCAAUU----------------------------- .....(((((.........(((((((........))))))).)))))....((((((((.........))))))))....----------------------------- ( -27.60, z-score = -2.65, R) >droEre2.scaffold_4820 7798238 109 - 10470090 AGUAAUGGGCUGCCGUUAAAGGUGGCGACCAGGAGCCGCCUAGCCCGACAAGUGAUUGUUUUAUAAUUGCAGUCGCAAUUGCAGCCUGCAGCUUGCCACAGGCUACUUG .((((((((((........(((((((........)))))))))))).....((((((((.........)))))))).)))))((((((..(....)..))))))..... ( -42.50, z-score = -2.10, R) >droYak2.chr3R 7683261 109 - 28832112 AGUAAUGGGCUGCCGUUAAAGGUGGCGACCAGGAGCCGCCUAUCCCGACAAGUGAUUGUUUUAUAAUUGCAAUCGCAAUUGCAACCUGCAGCUUGCCGCUUGCCACUUG ..((((((....)))))).(((((((........))))))).....(.(((((((((((.........))))))((((((((.....)))).)))).))))).)..... ( -35.30, z-score = -1.22, R) >droSec1.super_4 4191977 94 + 6179234 AGUAAUGGGUUGCCGUUAAAGGUGGCGACCGGGAGCCGCCUAUACCGACAAGUGAUUGUUUUAUAAUUGCAAUCGCAAUUGCAGCUUGCAGCUU--------------- (((..((((((((((.((.(((((((........))))))).)).))....((((((((.........))))))))....))))))))..))).--------------- ( -32.90, z-score = -2.03, R) >droSim1.chr3R 25006733 94 + 27517382 AGUAAUGGGCUGCCGUUAAAGGUGGCGACCGGGAGCCGCCUAUCCCGACAAGUGAUUGUUUUAUAAUUGCAAUCGCAAUUGCAGCUUGCAGCUU--------------- (((..((((((((((..(.(((((((........))))))).)..))....((((((((.........))))))))....))))))))..))).--------------- ( -34.50, z-score = -2.06, R) >droPer1.super_7 2364079 91 + 4445127 AAUAAUGGGGAGCCGUUAAAGGUGGCGACCGGGAGCCGCCUAGCCCGACAAGUGAUUGUUUUAUAAUUGCAAUCGCAAUUGCAGUUCGAUG------------------ ..((((((....)))))).(((((((........))))))).....(((((....)))))....(((((((((....))))))))).....------------------ ( -29.60, z-score = -1.67, R) >dp4.chr2 2196951 91 + 30794189 AAUAAUGGGGAGCCGUUAAAGGUGGCGACCGGGAGCCGCCUAGCCCGACAAGUGAUUGUUUUAUAAUUGCAAUCGCAAUUGCAGUUCGAUG------------------ ..((((((....)))))).(((((((........))))))).....(((((....)))))....(((((((((....))))))))).....------------------ ( -29.60, z-score = -1.67, R) >droWil1.scaffold_181089 5574787 91 + 12369635 AAUAAUGGUAAGUCGUUAAAGGUGGCGACCAGGAGCCGCCUAUCCGGACAAGUGAUUGUUUUAUAAUUGCAAUUGCAAUUGCAGUUCGAUG------------------ ..((((((....)))))).(((((((........)))))))...(((((..(..(((((...)))))..)...(((....))))))))...------------------ ( -27.30, z-score = -2.36, R) >droVir3.scaffold_12855 657814 88 + 10161210 -AUAAUGGGCAGCCGUUAA-GGUGGCGACCGGGAGCCGCCUAGCCGGACAAGUGAUUGUUUUAUAAUUGCAAUCGCAAUUGCAGUUCGAU------------------- -.((((((....))))))(-((((((........)))))))...(((((....((((((.........))))))((....)).)))))..------------------- ( -30.60, z-score = -1.90, R) >consensus AAUAAUGGGCAGCCGUUAAAGGUGGCGACCGGGAGCCGCCUAGCCCGACAAGUGAUUGUUUUAUAAUUGCAAUCGCAAUUGCAGCUUGCAG__________________ ..((((((....)))))).(((((((........)))))))..........((((((((.........))))))))................................. (-26.55 = -26.38 + -0.17)
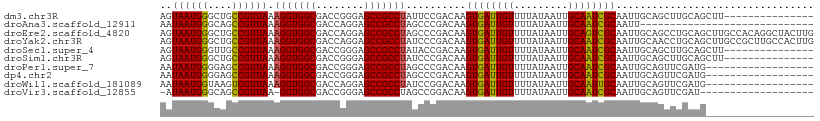
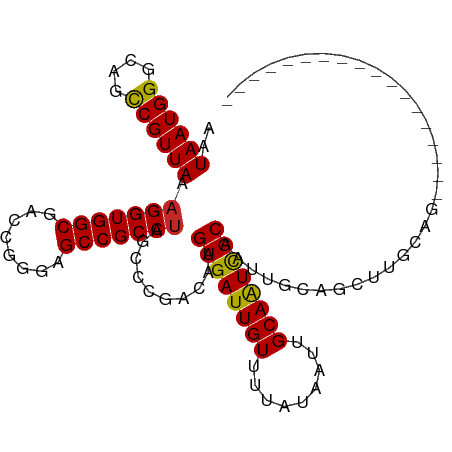
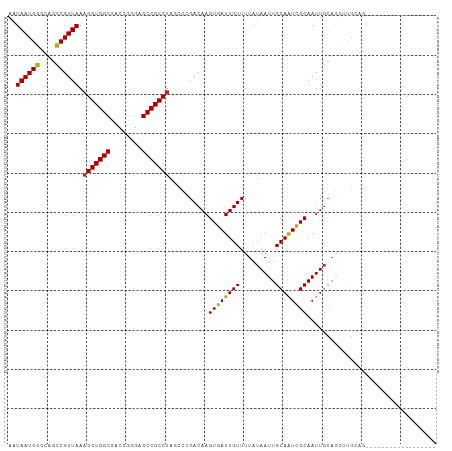
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:54:47 2011