| Sequence ID | dm3.chr2L |
|---|---|
| Location | 9,096,871 – 9,096,962 |
| Length | 91 |
| Max. P | 0.761273 |

| Location | 9,096,871 – 9,096,962 |
|---|---|
| Length | 91 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 68.84 |
| Shannon entropy | 0.62903 |
| G+C content | 0.37564 |
| Mean single sequence MFE | -16.35 |
| Consensus MFE | -4.62 |
| Energy contribution | -4.71 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.28 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.761273 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
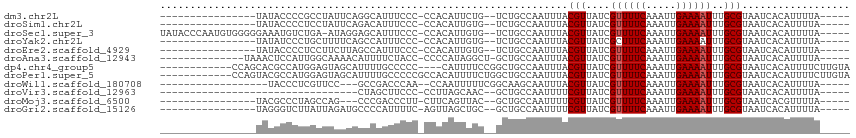
>dm3.chr2L 9096871 91 + 23011544 ----------------UAUACCCCGCCUAUUCAGGCAUUUCCC-CCACAUUCUG--UCUGCCAAUUUACGUUAUCGUUUUCAAAUUGAAAAUUUGCGUAAUCACAUUUUA----- ----------------........((((....)))).......-..........--.........((((((....((((((.....))))))..))))))..........----- ( -14.10, z-score = -2.90, R) >droSim1.chr2L 8877024 91 + 22036055 ----------------UAUACCCCUCCUAUUCAGACAUUUCCC-CCACAUUGUG--UCUGCCAAUUUACGUUAUCGUUUUCAAAUUGAAAAUUUGCGUAAUCACAUUUUA----- ----------------...............(((((((.....-.......)))--)))).....((((((....((((((.....))))))..))))))..........----- ( -15.50, z-score = -4.36, R) >droSec1.super_3 4563235 106 + 7220098 UAUACCCAAUGUGGGGGAAAUGUCUGA-AUAGGAGCAUUUCCC-CCACAUUGUG--UCUGCCAAUUUACGUUAUCGUUUUCAAAUUGAAAAUUUGCGUAAUCACAUUUUA----- ...((.(((((((((((((((((((..-...))).))))))))-)))))))).)--)........((((((....((((((.....))))))..))))))..........----- ( -39.30, z-score = -5.97, R) >droYak2.chr2L 11758171 91 + 22324452 ----------------UAUAUCCCUGCUUUUCAGCCAUUUCCC-CCACAUUGUG--UCUGCCAAUUUACGUUAUCGCUUUCAAAUUGAAAAUUUGCGUAAUCACAUUUUA----- ----------------.........(((....)))........-......((((--..((((((....((....)).((((.....))))..))).)))..)))).....----- ( -8.20, z-score = -0.42, R) >droEre2.scaffold_4929 9701392 91 + 26641161 ----------------UAUACCCCUCCUUCUUAGCCAUUUCCC-CCACAUUGUG--UCUGCCAAUUUACGUUAUCGUUUUCAAAUUGAAAAUUUGCGUAAUCACAUUUUA----- ----------------...........................-......((((--.........((((((....((((((.....))))))..)))))).)))).....----- ( -10.20, z-score = -2.41, R) >droAna3.scaffold_12943 443781 94 + 5039921 --------------UAAACUCCAUUGGCAAAACAUUUUCUACC-CCCCAUAGGCU-GCUGCCAAUUUACGUUAUCGUUUUCAAAUUGAAAAUUUGCGUAAUCACAUUUUA----- --------------........(((((((...((...((((..-.....)))).)-).)))))))((((((....((((((.....))))))..))))))..........----- ( -15.10, z-score = -1.57, R) >dp4.chr4_group5 1395776 99 + 2436548 ------------CCAGCACGCCAUGGAGUAGCAUUUUGCCCCC----CAUUUUCCGGCUGCCAAUUUACGUUAUCGUUUUCAAAUUGAAAAUUUGCGUAAUCACAUUUUCUUGUA ------------...(((.(((((((.(..((.....))..))----))).....))))))....((((((....((((((.....))))))..))))))..(((......))). ( -21.70, z-score = -2.24, R) >droPer1.super_5 5802011 103 + 6813705 ------------CCAGUACGCCAUGGAGUAGCAUUUUGCCCCCGCCACAUUUUCUGGCUGCCAAUUUACGUUAUCGUUUUCAAAUUGAAAAUUUGCGUAAUCACAUUUUCUUGUA ------------...(((.(((((((.(..((.....))..)..))).......)))))))....((((((....((((((.....))))))..))))))..(((......))). ( -18.51, z-score = -0.41, R) >droWil1.scaffold_180708 13917 87 + 12563649 ------------------UACCCUCGUUCC---GCCGACCCAA--CCAAUUUUUCGGCAAGCAAUUUACGUUAUCGUUUUCAAAUUGAAAAUUUGCGUAAUCACAUUUUA----- ------------------.......(((..---(((((.....--........))))).)))...((((((....((((((.....))))))..))))))..........----- ( -16.12, z-score = -2.60, R) >droVir3.scaffold_12963 430317 74 + 20206255 ---------------------------------CUAGCUUCCC-CCUUAGCAAC--GCUGCCAAUUUUCGUUAUCGUUUUCAAAUUGAAAAUUUGCGUAAUCACAUUUUA----- ---------------------------------...(((....-....))).((--((....((((((((...............)))))))).))))............----- ( -10.46, z-score = -1.81, R) >droMoj3.scaffold_6500 11927886 88 + 32352404 ----------------UACGCCCUAGCCAG---CCCGACCCUU-CUUCAGUUAC--GCUGCCAAUUUUCGUUAUCGUUUUCAAAUUGAAAAUUUGCGUAAUCACGUUUUA----- ----------------(((((....(.(((---(..(((....-.....)))..--)))).).............((((((.....))))))..)))))...........----- ( -13.40, z-score = -1.32, R) >droGri2.scaffold_15126 5214711 91 - 8399593 ----------------UAGGGUCUUAUUAGAUGCCCCAUUUUC-AGUUAGCUGC--GCUGCCAAUUUUCGUUAUCGUUUUCAAAUUGAAAAUUUGCGUAAUCACAUUUUA----- ----------------..((((.((....)).)))).......-.......(((--((....((((((((...............)))))))).)))))...........----- ( -13.66, z-score = 0.06, R) >consensus ________________UAUACCCUUGCUAUUCAGCCAUUUCCC_CCACAUUGUC__GCUGCCAAUUUACGUUAUCGUUUUCAAAUUGAAAAUUUGCGUAAUCACAUUUUA_____ ....................................................................(((....((((((.....))))))..))).................. ( -4.62 = -4.71 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:27:33 2011