
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 25,317,732 – 25,317,835 |
| Length | 103 |
| Max. P | 0.520320 |

| Location | 25,317,732 – 25,317,835 |
|---|---|
| Length | 103 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.92 |
| Shannon entropy | 0.47481 |
| G+C content | 0.51530 |
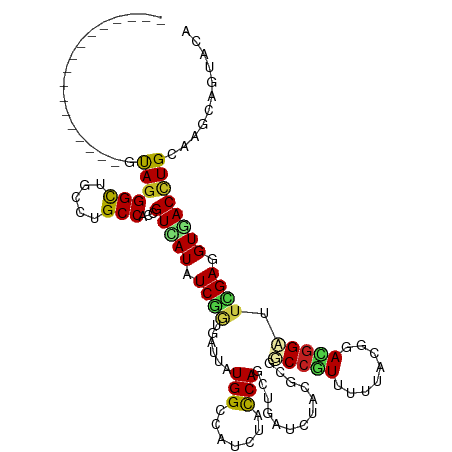
| Mean single sequence MFE | -34.25 |
| Consensus MFE | -15.45 |
| Energy contribution | -15.20 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.52 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.520320 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
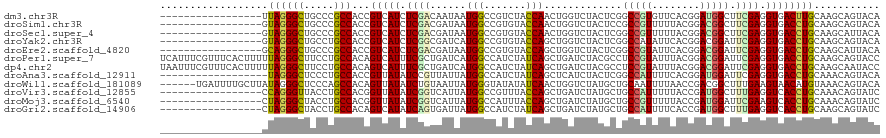
>dm3.chr3R 25317732 103 - 27905053 -----------------UUAGGGCUGCCCGCCACCGUCAUCUCGACAAUAAUGGCCGUCUACCAACUGGUCUACUCGGCCGUGUUCACGGAUGGCUUCGAGGUGACUUGCAAGCAGUACA -----------------...(..((((..((....((((((((((.......((((((((...((((((((.....))))).)))...))))))))))))))))))..))..))))..). ( -42.21, z-score = -3.21, R) >droSim1.chr3R 24990515 103 - 27517382 -----------------GUAGGGCUGCCCGCCACCGUCAUCUCGACGAUAAUGGCCGUGUACCAACUGGUCUACUCCGCCGUUUUUACGGACGGCUUCGAGGUGACCUGCAAGCAGUACA -----------------...(..((((..((....((((((((((.......((..(((.(((....))).))).))((((((......)))))).))))))))))..))..))))..). ( -40.10, z-score = -2.27, R) >droSec1.super_4 4176229 103 - 6179234 -----------------GUAGGGCUGCCCGCCACCGUCAUCUCGACGAUAAUGGCCGUGUACCAACUGGUCUACUCGGCCGUUUUUACGGACGGCUUCGAGGUGACCUGCAAGCAUUACA -----------------(((((((.....)))...(((((((((((((....(((((.((....)))))))...)))((((((......)))))).)))))))))))))).......... ( -41.10, z-score = -2.73, R) >droYak2.chr3R 7666720 103 + 28832112 -----------------GUAGGGCUGCCUGCCACCGUCAUCUCGGCGAUCAUGGCCGUGUACCAGCUGGUCUACUCGGCCAUAUUCACGGACGGAUUCGAGGUGACCUGCAAGCAGUACA -----------------...(..((((.(((....((((((((((.((..(((((((.(((((....)))..)).)))))))..)).(....)...))))))))))..))).))))..). ( -43.30, z-score = -2.31, R) >droEre2.scaffold_4820 7782385 103 + 10470090 -----------------GCAGGGCUGCCCGCCACCGUCAUCUCGACGAUAAUGGCCGUGUACCAGCUGGUCUACUCGGCCGUAUUCACGGACGGAUUCGAGGUGACCUGCAAGCAUUACA -----------------(((((((.....)))...((((((((((.....(((((((.(((((....)))..)).)))))))((((......)))))))))))))))))).......... ( -42.00, z-score = -2.45, R) >droPer1.super_7 2347045 120 - 4445127 UCAUUUCGUUUCACUUUUUAGGGCUUCCUGCCACAGUCAUUUCGCUGAUCAUGGCCAUCUAUCAGCUGAUCUACGCCUCCGUAUUUACGGACGGAUUCGAGGUGACCUGCAAGCAGUACC ((((((((..((.........(((.....((((..((((......))))..)))).(((........)))....)))(((((....)))))..))..)))))))).(((....))).... ( -31.40, z-score = -1.17, R) >dp4.chr2 2179548 120 - 30794189 UAAUUUCGUUUCACUUUUUAGGGCUUCCUGCCACAGUCAUUUCGCUGAUCAUGGCCAUCUAUCAGCUGAUCUACGCCUCCGUAUUUACGGACGGAUUCGAGGUGACCUGCAAGCAAUACC .......((((((........(((.....)))...((((((((((((((...........)))))).........(((((((....))))).))....)))))))).)).))))...... ( -31.10, z-score = -1.49, R) >droAna3.scaffold_12911 5006309 102 - 5364042 ------------------UAGGGCUCCCUGCCACCGUUAUAUCCGUUAUUAUGGCCAUCUAUCAGCUCAUCUACUCGGCCAUUUUCACGGAUGGAUUCGAGGUGACCUGCAAACAGUACA ------------------((((....)))).((((....(((((((.(..((((((....................))))))..).))))))).......))))..(((....))).... ( -28.05, z-score = -1.23, R) >droWil1.scaffold_181089 5558929 114 - 12369635 ------UGAUUUUGCUUAUAGGGCUCCCAGCCACAGUUAUAUCUGUAAUUAUGGGUAUAUAUCAACUGGUCUAUGCUGCAAUUUUAACCGACGGCUUUGAAGUAACAUGUAAACAGUACA ------((((..((((((((((((.....)))((((......))))..)))))))))...))))((((.....(((((..(((((((((...))..)))))))..)).)))..))))... ( -23.20, z-score = 0.49, R) >droVir3.scaffold_12855 640929 103 - 10161210 -----------------CCAGGGUUACCUGCCACGGUUAUAUCGGUCAUUAUGGCCGUUUACCAGCUGAUCUAUGCUGCCAUUUUUACCGAUGGCUUUGAGGUCACCUGCAAACAGUAUC -----------------.((((....))))..(((((((((........)))))))))....(((.((((((.....((((((......))))))....)))))).)))........... ( -32.10, z-score = -1.91, R) >droMoj3.scaffold_6540 21818520 103 + 34148556 -----------------CUAGGGCUACCUGCCACGGUUAUAUCGGUCAUUAUGGCCAUUUACCAGCUGAUCUAUGCUGCCGUUUUUACCGAUGGAUUCGAAGUCACCUGCAAACAGUAUC -----------------....(((.....))).(((...(((((((.(..(((((........(((........))))))))..).)))))))...))).......(((....))).... ( -25.40, z-score = -0.37, R) >droGri2.scaffold_14906 1750276 103 - 14172833 -----------------CUAGGGCUACCUGCCACAGUCAUAUCAGUGAUUAUGGCCAUCUAUCAGCUGAUCUAUGCUGCCAUUUUCACCGAUGGCUUUGAGGUCACCUGCAAGCAGUAUC -----------------.....(((....((((.((((((....)))))).)))).......(((.((((((.....((((((......))))))....)))))).)))..)))...... ( -31.00, z-score = -1.06, R) >consensus _________________GUAGGGCUGCCUGCCACCGUCAUAUCGGUGAUUAUGGCCAUCUACCAGCUGAUCUACGCGGCCGUUUUUACGGACGGAUUCGAGGUGACCUGCAAGCAGUACA ..................((((((.....)))...(((((.((((......(((.......))).............(((((........))))).)))).))))))))........... (-15.45 = -15.20 + -0.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:54:43 2011