
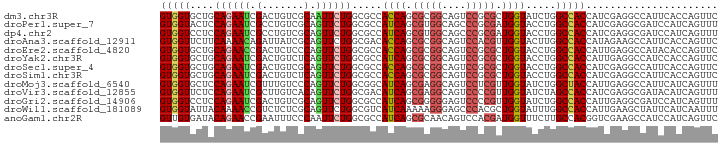
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 25,315,905 – 25,315,998 |
| Length | 93 |
| Max. P | 0.711547 |

| Location | 25,315,905 – 25,315,998 |
|---|---|
| Length | 93 |
| Sequences | 13 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 83.82 |
| Shannon entropy | 0.36895 |
| G+C content | 0.58313 |
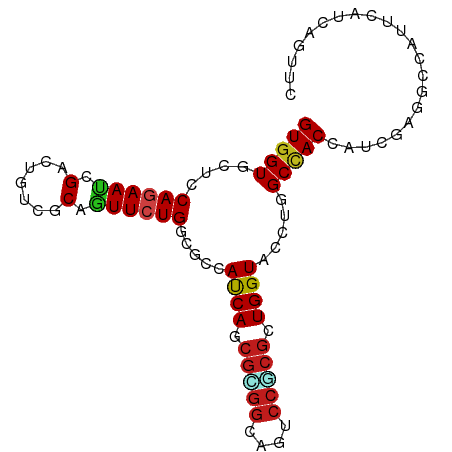
| Mean single sequence MFE | -34.28 |
| Consensus MFE | -19.99 |
| Energy contribution | -20.15 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.711547 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 25315905 93 + 27905053 GUGGUGCUGCAGAAUCGACUGUCGCAAUUCUGGCGCCACCAGCGCGGCAGUCCGCGCUGGUAUCUGGCCACCAUCGAGGCCAUUCACCAGUUC ..(((((..((((((((.....))..)))))))))))((((((((((....))))))))))...(((((........)))))........... ( -41.90, z-score = -2.93, R) >droPer1.super_7 2344820 93 + 4445127 GUGGUACUCCAGAAUCGCCUGUCGCAGUUCUGGCGCCAUCAGCGUGGCAGCCCGCGAUGGUACCUGGCCACCAUCGAGGCGAUCCAUCAGUUU ((((....(((((((.((.....)).))))))).(((((....))))).(((..(((((((........))))))).)))...))))...... ( -34.30, z-score = -1.06, R) >dp4.chr2 2177179 93 + 30794189 GUGGUCCUCCAGAAUCGCCUGUCGCAGUUCUGGCGCCAUCAGCGUGGCAGCCCGCGAUGGUACCUGGCCACCAUCGAGGCGAUCCAUCAGUUU ((((((..(((((((.((.....)).))))))).(((((....))))).(((..(((((((........))))))).))))).))))...... ( -35.50, z-score = -1.38, R) >droAna3.scaffold_12911 5003917 93 + 5364042 GUGGUUCUUCAAAACAGAUUAUCGCAGUUCUGGCGACACCAGCGCGGCAGUCCACGUUGGUACUUGGCCACCAUAGAAGCCAUUCACCAGUUC (((((...((......))..)))))....((((.((.(((((((.((....)).)))))))...((((..........)))).)).))))... ( -27.10, z-score = -1.79, R) >droEre2.scaffold_4820 7780259 93 - 10470090 GUGGUGCUGCAGAACCGACUCUCCCAGUUCUGGCGCCACCAGCGCGGCAGUCCGCGCUGGUACCUGGCCACCAUUGAGGCCAUACACCAGUUC (((((((..((((((.(.......).)))))))))))))((((((((....))))))))(((..(((((........))))))))........ ( -42.80, z-score = -3.19, R) >droYak2.chr3R 7664089 93 - 28832112 GUGGUGCUGCAGAAUCGACUGUCUCAGUUCUGGCGCCAUCAGCGCGGCAGUCCGCGCUGGUACCUGGCCACCAUUGAGGCCAUCCACCAGUUC ((((.((((((((((.((.....)).))))).((((.....)))).)))))))))((((((...(((((........)))))...)))))).. ( -40.00, z-score = -2.22, R) >droSec1.super_4 4174333 93 + 6179234 GUGGUGCUGCAGAAUCGACUGUCGCAGUUCUGGCGCCACCAGCGCGGCAGUCCGCGCUGGUACCUGGCCACCAUCGAGGCCAUUCACCAGUUC ..(((((..((((((((.....))..)))))))))))((((((((((....))))))))))...(((((........)))))........... ( -42.20, z-score = -2.50, R) >droSim1.chr3R 24988615 93 + 27517382 GUGGUGCUGCAGAAUCGACUGUCUCAGUUCUGGCGCCACCAGCGCGGCAGUCCGCGCUGGUACCUGGCCACCAUCGAGGCCAUUCACCAGUUC ..(((((..((((((.((.....)).)))))))))))((((((((((....))))))))))...(((((........)))))........... ( -42.50, z-score = -2.99, R) >droMoj3.scaffold_6540 21816813 93 - 34148556 GUGGUGCUCCAGAAUCGUUUGUCCCAGUUCUGGCGGCAUCAGCGAGGCAGUCCUCGUUGGUAUCUGGCUACCAUUGAGGCCAUUCAUCAGUUU ((((((..(((((.......((((((....))).)))((((((((((....)))))))))).))))).))))))((((....))))....... ( -33.10, z-score = -2.22, R) >droVir3.scaffold_12855 639277 93 + 10161210 GUGGUUCUCCAGAAUCGCUUGUCACAGUUCUGGCGACAUCAGCGAGGCAGUCCCCGUUGGUAUCUAGCCACCAUCGAGGCGAUACAUCAGUUU (((((((....))))))).((((.((....))..)))).(((((.((.....)))))))(((((..(((........))))))))........ ( -26.90, z-score = -1.01, R) >droGri2.scaffold_14906 1748052 93 + 14172833 GUGGUCCUCCAGAAUCGACUGUCGCAGUUCUGGCGCCAUCAGCGGGGGAGUCCCCGUUGGUAUCUGGCCACCAUUGAGGCGAUCCAUCAGUUU (((((((.(((((((((.....))..))))))).)..((((((((((....))))))))))....)))))).(((((((....)).))))).. ( -36.50, z-score = -1.63, R) >droWil1.scaffold_181089 5557426 93 + 12369635 GUGGUAUUACAAAACCGUCUCUCGCAGUUCUGGCGUCAUCAAAAAGGGAGCCCACGCUGGUAUUUGGCCACCAUUGAAGCUAUUCAUCAAUUU (((((..(((.....((.....))((((...(((.((.((.....)))))))...)))))))....)))))...((((....))))....... ( -17.70, z-score = 0.74, R) >anoGam1.chr2R 55954533 93 + 62725911 GUUGUGAUACAGAACCGAAUUUCCCAAUUCUGGCGCCAUCAGCGCAACAGUCCACGAUGGUUUCUUGCCACGGUCGAAGCCAUCCAUCAGUUC ...(((...(((((..(.......)..)))))((((.....)))).......)))(((((((((..((....)).)))))))))......... ( -25.10, z-score = -1.67, R) >consensus GUGGUGCUCCAGAAUCGACUGUCGCAGUUCUGGCGCCAUCAGCGCGGCAGUCCGCGCUGGUACCUGGCCACCAUCGAGGCCAUUCAUCAGUUC (((((....((((((.(.......).)))))).....((((.(((((....))))).)))).....)))))...................... (-19.99 = -20.15 + 0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:54:42 2011