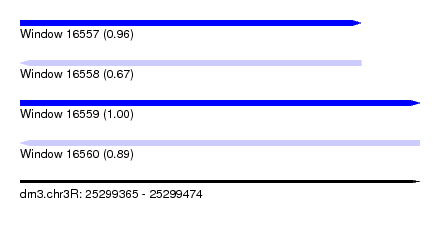
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 25,299,365 – 25,299,474 |
| Length | 109 |
| Max. P | 0.996839 |

| Location | 25,299,365 – 25,299,458 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 78.67 |
| Shannon entropy | 0.39512 |
| G+C content | 0.52835 |
| Mean single sequence MFE | -27.48 |
| Consensus MFE | -21.09 |
| Energy contribution | -21.87 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.956723 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
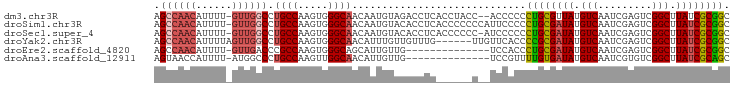
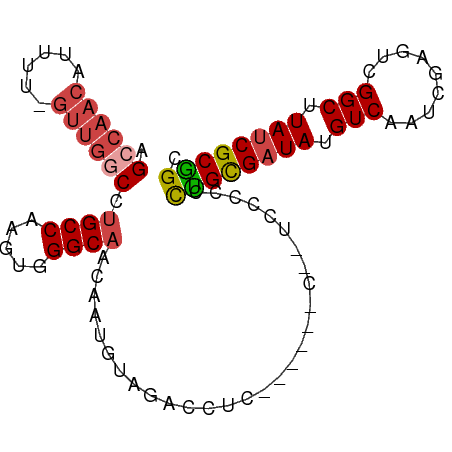
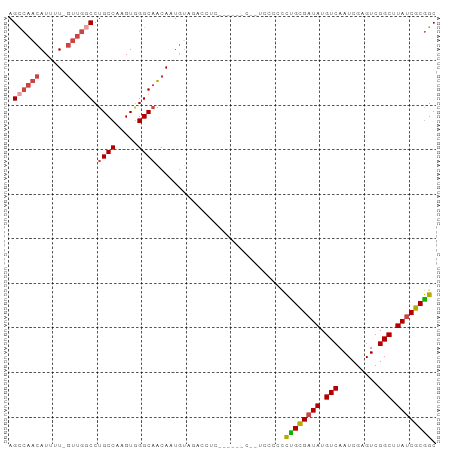
>dm3.chr3R 25299365 93 + 27905053 AGCCAACAUUUU-GUUGGCCUGCCAAGUGGGCAACAAUGUAGACCUCACCUACC--ACCCCCCUGCGUUAUGUCAAUCGAGUCGGCUUAUCGCGGC .((((((.....-))))))..(((.((.(((.......((((.......)))).--...)))))(((.((.(((.........))).)).)))))) ( -24.80, z-score = -0.27, R) >droSim1.chr3R 24972660 95 + 27517382 AGCCAACAUUUU-GUUGGCCUGCCAAGUGGGCAACAAUGUACACCUCACCCCCCCAUUCCCCCUGCGAUAUGUCAAUCGAGUCGGCUUAUCGCGGC .((((((.....-))))))..(((.((((((......((.......))....))))))......((((((.(((.........))).))))))))) ( -27.60, z-score = -1.51, R) >droSec1.super_4 4157966 94 + 6179234 AGCCAACAUUUU-GUUGGCCUGCCAAGUGGGCAACAAUGUACACCUCACCCCCC-AUCCCCCCUGCGAUAUGUCAAUCGAGUCGGCUUAUCGCGGC .((((((.....-))))))..(((..(((..((....))..)))..........-.........((((((.(((.........))).))))))))) ( -27.50, z-score = -1.39, R) >droYak2.chr3R 7647078 90 - 28832112 AGCCAACAUUUUAGUUGGCCUGCCAAGUGGGCAACAUUUGUUGUUUG------UUGUUCACCCCGCGAUAUGUCAAUCGAGUCGGCUUAUCGCGGC .((((((......)))))).......((((((((((.........))------)))))))).((((((((.(((.........))).)))))))). ( -35.80, z-score = -4.07, R) >droEre2.scaffold_4820 7763234 81 - 10470090 AGCCAACAUUUU-GUUGACCCGCCAAGUGGGCAGCAUUGUUG--------------UCCACCCUGCGAUAUGUCAAUCGAGUCGGCUUAUCGCGGC ...(((((...(-((((.(((((...)))))))))).)))))--------------......((((((((.(((.........))).)))))))). ( -30.00, z-score = -2.65, R) >droAna3.scaffold_12911 4987212 81 + 5364042 AGUAACCAUUUU-AUGGCCCUGCCAAGUUGGCAACAUUGUUG--------------UCCGUUUUGUGAUAUGUCAAUCGUGUCGGCUUAUCGCAGC .....((((...-))))..((((.(((((((((..((((.((--------------((........))))...))))..)))))))))...)))). ( -19.20, z-score = -0.51, R) >consensus AGCCAACAUUUU_GUUGGCCUGCCAAGUGGGCAACAAUGUAGACCUC______C__UCCCCCCUGCGAUAUGUCAAUCGAGUCGGCUUAUCGCGGC .((((((......)))))).((((.....)))).............................((((((((.(((.........))).)))))))). (-21.09 = -21.87 + 0.78)
| Location | 25,299,365 – 25,299,458 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 78.67 |
| Shannon entropy | 0.39512 |
| G+C content | 0.52835 |
| Mean single sequence MFE | -29.35 |
| Consensus MFE | -18.28 |
| Energy contribution | -19.03 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.666715 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 25299365 93 - 27905053 GCCGCGAUAAGCCGACUCGAUUGACAUAACGCAGGGGGGU--GGUAGGUGAGGUCUACAUUGUUGCCCACUUGGCAGGCCAAC-AAAAUGUUGGCU ((((((.((.(.(((.....))).).)).)))(((.((((--..((((((.....))).)))..)))).)))))).(((((((-.....))))))) ( -30.30, z-score = -0.35, R) >droSim1.chr3R 24972660 95 - 27517382 GCCGCGAUAAGCCGACUCGAUUGACAUAUCGCAGGGGGAAUGGGGGGGUGAGGUGUACAUUGUUGCCCACUUGGCAGGCCAAC-AAAAUGUUGGCU (((((((((.(.(((.....))).).))))))..(((.((..(....(((.....))).)..)).)))....))).(((((((-.....))))))) ( -32.60, z-score = -0.77, R) >droSec1.super_4 4157966 94 - 6179234 GCCGCGAUAAGCCGACUCGAUUGACAUAUCGCAGGGGGGAU-GGGGGGUGAGGUGUACAUUGUUGCCCACUUGGCAGGCCAAC-AAAAUGUUGGCU (((((((((.(.(((.....))).).))))))..(((.((.-.(...(((.....))).)..)).)))....))).(((((((-.....))))))) ( -33.60, z-score = -1.07, R) >droYak2.chr3R 7647078 90 + 28832112 GCCGCGAUAAGCCGACUCGAUUGACAUAUCGCGGGGUGAACAA------CAAACAACAAAUGUUGCCCACUUGGCAGGCCAACUAAAAUGUUGGCU (((((((((.(.(((.....))).).)))))).(((((..(((------((.........)))))..)))))))).(((((((......))))))) ( -32.00, z-score = -3.48, R) >droEre2.scaffold_4820 7763234 81 + 10470090 GCCGCGAUAAGCCGACUCGAUUGACAUAUCGCAGGGUGGA--------------CAACAAUGCUGCCCACUUGGCGGGUCAAC-AAAAUGUUGGCU (((((((((.(.(((.....))).).)))))).(((..(.--------------((....)))..)))....))).(((((((-.....))))))) ( -30.50, z-score = -2.60, R) >droAna3.scaffold_12911 4987212 81 - 5364042 GCUGCGAUAAGCCGACACGAUUGACAUAUCACAAAACGGA--------------CAACAAUGUUGCCAACUUGGCAGGGCCAU-AAAAUGGUUACU .((((((((.(.(((.....))).).)))).(((...((.--------------((((...))))))...)))))))((((((-...))))))... ( -17.10, z-score = -0.77, R) >consensus GCCGCGAUAAGCCGACUCGAUUGACAUAUCGCAGGGGGGA__G______GAGGUCAACAAUGUUGCCCACUUGGCAGGCCAAC_AAAAUGUUGGCU (((((((((.(.(((.....))).).)))))).(((.((.......................)).)))....))).(((((((......))))))) (-18.28 = -19.03 + 0.75)

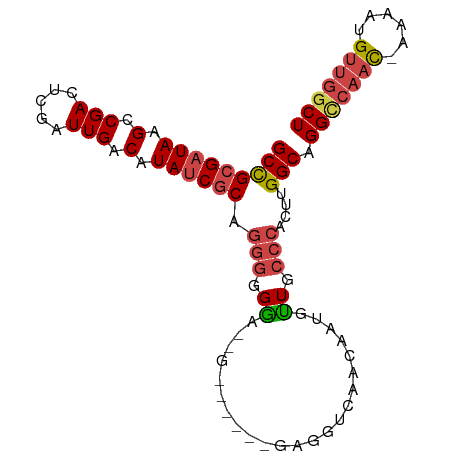
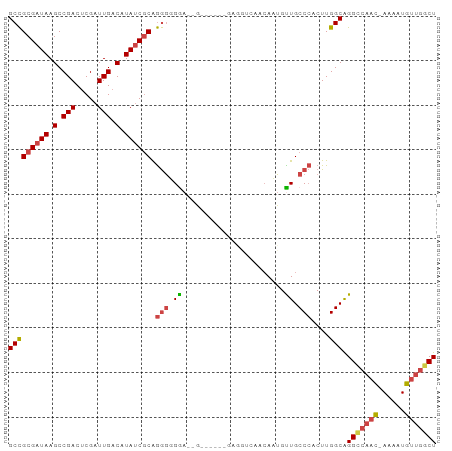
| Location | 25,299,365 – 25,299,474 |
|---|---|
| Length | 109 |
| Sequences | 7 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 70.79 |
| Shannon entropy | 0.55977 |
| G+C content | 0.53883 |
| Mean single sequence MFE | -34.99 |
| Consensus MFE | -18.45 |
| Energy contribution | -19.70 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.99 |
| SVM RNA-class probability | 0.996839 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 25299365 109 + 27905053 AGCCAACAUUUU-GUUGGCCUGCCAAGUGGGCAACAAU-GUAGACCUCACCUACC--ACCCCCCUGCGUUAUGUCAAUCGAGUCGGCUUAUCGCGGCCAUUU-GGCCCGUUCUG .((((((.....-))))))..(((((((((.(......-((((.......)))).--........(((.((.(((.........))).)).)))).))))))-)))........ ( -34.30, z-score = -1.82, R) >droSim1.chr3R 24972660 111 + 27517382 AGCCAACAUUUU-GUUGGCCUGCCAAGUGGGCAACAAU-GUACACCUCACCCCCCCAUUCCCCCUGCGAUAUGUCAAUCGAGUCGGCUUAUCGCGGCCAUUU-GGCCCGUUCUG .((((((.....-))))))..((((((((((((....)-))......................((((((((.(((.........))).))))))))))))))-)))........ ( -36.60, z-score = -2.81, R) >droSec1.super_4 4157966 110 + 6179234 AGCCAACAUUUU-GUUGGCCUGCCAAGUGGGCAACAAU-GUACACCUCACCCCCC-AUCCCCCCUGCGAUAUGUCAAUCGAGUCGGCUUAUCGCGGCCAUUU-GGCCCGUUCUG .((((((.....-))))))..((((((((((((....)-))..............-.......((((((((.(((.........))).))))))))))))))-)))........ ( -36.60, z-score = -2.83, R) >droYak2.chr3R 7647078 106 - 28832112 AGCCAACAUUUUAGUUGGCCUGCCAAGUGGGCAACAUUUGUUG-------UUUGUUGUUCACCCCGCGAUAUGUCAAUCGAGUCGGCUUAUCGCGGCCGUUU-GGCCCGGUCUG .((((((......))))))..(((((((((((((((.......-------..)))))))))).((((((((.(((.........))).))))))))....))-)))........ ( -42.20, z-score = -3.64, R) >droEre2.scaffold_4820 7763234 97 - 10470090 AGCCAACAUUUU-GUUGACCCGCCAAGUGGGCAGCAUU-GUUG--------------UCCACCCUGCGAUAUGUCAAUCGAGUCGGCUUAUCGCGGCCAUUU-GGCCCGGUCUG ((((...((((.-((((((.(((...(((((((((...-))))--------------)))))...)))....)))))).)))).)))).((((.((((....-))))))))... ( -38.00, z-score = -3.02, R) >droAna3.scaffold_12911 4987212 98 + 5364042 AGUAACCAUUUU-AUGGCCCUGCCAAGUUGGCAACAUU-GUUG--------------UCCGUUUUGUGAUAUGUCAAUCGUGUCGGCUUAUCGCAGCCAUUUUGGCCCGGCCCA ............-..((((..((((((.((((.(((..-..))--------------)......(((((((.(((.........))).))))))))))).))))))..)))).. ( -33.00, z-score = -2.77, R) >droPer1.super_7 2326894 82 + 4445127 --------------------UGCCACUUUUGUUGGACCUUUUC-----------UGGGGUGCCUUGUGAUGUGUCAAUCGUGCUGGCUUAUCGCGGCCAUUU-GGCUGGGAACA --------------------.((((........(((((((...-----------.))))).))....(((......)))....))))...((.(((((....-))))).))... ( -24.20, z-score = -0.17, R) >consensus AGCCAACAUUUU_GUUGGCCUGCCAAGUGGGCAACAAU_GUUG___________C__UCCCCCCUGCGAUAUGUCAAUCGAGUCGGCUUAUCGCGGCCAUUU_GGCCCGGUCUG .............((((.(((((...)))))))))..............................((((((.(((.........))).))))))((((.....))))....... (-18.45 = -19.70 + 1.25)

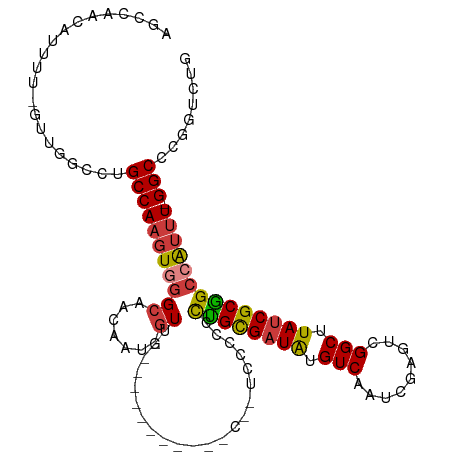
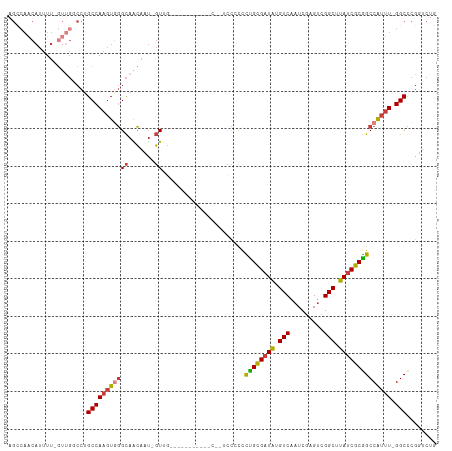
| Location | 25,299,365 – 25,299,474 |
|---|---|
| Length | 109 |
| Sequences | 7 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 70.79 |
| Shannon entropy | 0.55977 |
| G+C content | 0.53883 |
| Mean single sequence MFE | -33.59 |
| Consensus MFE | -17.67 |
| Energy contribution | -19.01 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.888588 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 25299365 109 - 27905053 CAGAACGGGCC-AAAUGGCCGCGAUAAGCCGACUCGAUUGACAUAACGCAGGGGGGU--GGUAGGUGAGGUCUAC-AUUGUUGCCCACUUGGCAGGCCAAC-AAAAUGUUGGCU .....((((((-....)))).))....((((((((...............)))((((--..((((((.....)))-.)))..))))..))))).(((((((-.....))))))) ( -37.16, z-score = -0.78, R) >droSim1.chr3R 24972660 111 - 27517382 CAGAACGGGCC-AAAUGGCCGCGAUAAGCCGACUCGAUUGACAUAUCGCAGGGGGAAUGGGGGGGUGAGGUGUAC-AUUGUUGCCCACUUGGCAGGCCAAC-AAAAUGUUGGCU .....((((((-....)))).))....(((((((((((......)))).))(((.((..(....(((.....)))-.)..)).)))..))))).(((((((-.....))))))) ( -39.10, z-score = -1.07, R) >droSec1.super_4 4157966 110 - 6179234 CAGAACGGGCC-AAAUGGCCGCGAUAAGCCGACUCGAUUGACAUAUCGCAGGGGGGAU-GGGGGGUGAGGUGUAC-AUUGUUGCCCACUUGGCAGGCCAAC-AAAAUGUUGGCU .....((((((-....)))).))....(((((((((((......)))).))(((.((.-.(...(((.....)))-.)..)).)))..))))).(((((((-.....))))))) ( -40.10, z-score = -1.37, R) >droYak2.chr3R 7647078 106 + 28832112 CAGACCGGGCC-AAACGGCCGCGAUAAGCCGACUCGAUUGACAUAUCGCGGGGUGAACAACAAA-------CAACAAAUGUUGCCCACUUGGCAGGCCAACUAAAAUGUUGGCU ........(((-(.....((((((((.(.(((.....))).).))))))))((((..(((((..-------.......)))))..)))))))).(((((((......))))))) ( -37.00, z-score = -2.70, R) >droEre2.scaffold_4820 7763234 97 + 10470090 CAGACCGGGCC-AAAUGGCCGCGAUAAGCCGACUCGAUUGACAUAUCGCAGGGUGGA--------------CAAC-AAUGCUGCCCACUUGGCGGGUCAAC-AAAAUGUUGGCU .....((((((-....)))).))...(((((((....(((((...((((..(((((.--------------((..-.....)).)))))..))))))))).-.....))))))) ( -36.70, z-score = -2.30, R) >droAna3.scaffold_12911 4987212 98 - 5364042 UGGGCCGGGCCAAAAUGGCUGCGAUAAGCCGACACGAUUGACAUAUCACAAAACGGA--------------CAAC-AAUGUUGCCAACUUGGCAGGGCCAU-AAAAUGGUUACU ..((((..(((((..(((((......)))))....(((......))).......((.--------------((((-...))))))...)))))..))))..-............ ( -28.90, z-score = -1.58, R) >droPer1.super_7 2326894 82 - 4445127 UGUUCCCAGCC-AAAUGGCCGCGAUAAGCCAGCACGAUUGACACAUCACAAGGCACCCCA-----------GAAAAGGUCCAACAAAAGUGGCA-------------------- ...((.(.(((-....))).).))...((((....(((......)))....((.(((...-----------.....)))))........)))).-------------------- ( -16.20, z-score = 0.19, R) >consensus CAGACCGGGCC_AAAUGGCCGCGAUAAGCCGACUCGAUUGACAUAUCGCAGGGGGGA__G___________CAAC_AAUGUUGCCCACUUGGCAGGCCAAC_AAAAUGUUGGCU .......((((.....))))((((((.(.(((.....))).).))))))...............................(((((.....)))))((((((......)))))). (-17.67 = -19.01 + 1.34)

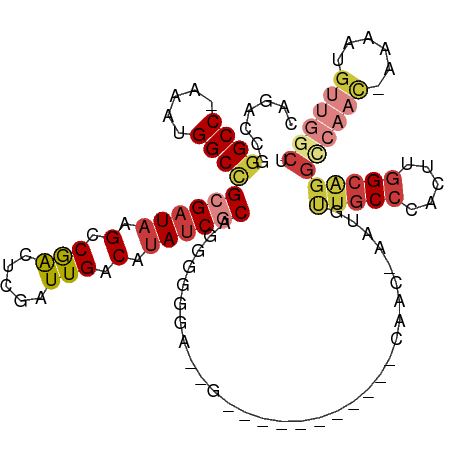
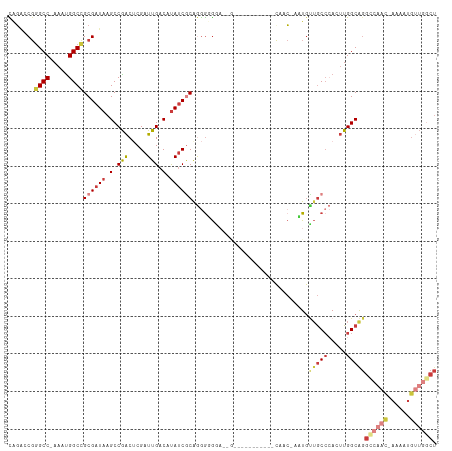
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:54:39 2011