| Sequence ID | dm3.chr2L |
|---|---|
| Location | 9,076,392 – 9,076,492 |
| Length | 100 |
| Max. P | 0.861118 |

| Location | 9,076,392 – 9,076,492 |
|---|---|
| Length | 100 |
| Sequences | 7 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 74.30 |
| Shannon entropy | 0.50479 |
| G+C content | 0.37241 |
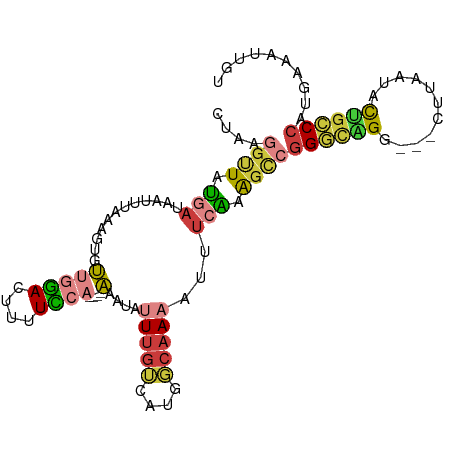
| Mean single sequence MFE | -25.86 |
| Consensus MFE | -12.24 |
| Energy contribution | -12.92 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.56 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.861118 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
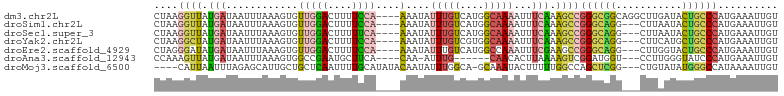
>dm3.chr2L 9076392 100 + 23011544 CUAAGGUUAUGAUAAUUUAAAGUGUUGGACUUUUCCA----AAAUAUUUGUCAUGGCAAAAUUUCAAAGCCGGGCGGCAGGCUUGAUACUGCCCAUGAAAUUGU .....((((((((((.........(((((....))))----).....))))))))))..(((((((..(((....))).(((........)))..))))))).. ( -27.34, z-score = -1.80, R) >droSim1.chr2L 8856524 97 + 22036055 CUAAGGUUAUGAUAAUUUAAAGUGUUGGACUUUUCCA----AAAUAUUUGUCAUGGCAAAAUUUCAAAGCCGGGCAGG---CUUAAUACUGCCCAUGAAAUUGU .....((((((((((.........(((((....))))----).....))))))))))..(((((((.....((((((.---.......)))))).))))))).. ( -28.54, z-score = -3.17, R) >droSec1.super_3 4542423 97 + 7220098 CUAAGGUUAUGAUAAUUUAAAGUGUUGGACUUUUUCA----AAAUAUUUGUCAUGGCAAAAUUUCAAAGCCGGGCAGG---CUUAAUACUGCCCAUGAAAUUGU .....((((((((((.........(((((....))))----).....))))))))))..(((((((.....((((((.---.......)))))).))))))).. ( -25.64, z-score = -2.42, R) >droYak2.chr2L 11737198 97 + 22324452 CUAAGGCUAUGAUAAUUUAAAGUGUUGGACUUUUCCA----AAAUAUUUGUCGUGGCAAAAUUUCAAAGCCGGGCAGG---CUUCAUGCUGCCCAUGAAAUUGU .....((((((((((.........(((((....))))----).....))))))))))..(((((((.....((((((.---(.....))))))).))))))).. ( -29.84, z-score = -2.76, R) >droEre2.scaffold_4929 9677219 97 + 26641161 CUAGGGAUAUGAUAAUUUAAAGUGUUGGACUUUUCCA----AAAUAUUUGUCAUGGCCAAAUUUCGAAGCCGGGCAGG---CUUGGUACUGCCCAUGAAAUUGU ....((.((((((((.........(((((....))))----).....)))))))).)).(((((((....)((((((.---.......))))))..)))))).. ( -27.34, z-score = -1.62, R) >droAna3.scaffold_12943 415306 90 + 5039921 CCAAAGUUAUGAUAAUUUAAAGUGGCCGAAUGCUUCA----CAA-AUUUG------CAACACUUAAAAGUCGGAUGGU---CCUUGGGUAUCCCAUGAAAUUGU .(((..(((((....(((.(((((..(((((......----...-)))))------...))))).)))...(((((.(---.....).))))))))))..))). ( -16.60, z-score = 0.19, R) >droMoj3.scaffold_6500 11899302 96 + 32352404 ----CAUUAAUUUAGAGCAUUGCUGCUCAAUUUUGCAUAUACAAUAUUUGGCA-GCAAAUACUUUUUGGCCAGCUCGG---CUGUAUAUGGGCCAUAAAAUUGU ----...((((((.((((.((((((((.(((.(((......))).))).))))-))))...(.....)....))))((---((.......))))...)))))). ( -25.70, z-score = -1.31, R) >consensus CUAAGGUUAUGAUAAUUUAAAGUGUUGGACUUUUCCA____AAAUAUUUGUCAUGGCAAAAUUUCAAAGCCGGGCAGG___CUUAAUACUGCCCAUGAAAUUGU .......((((((((..........((((....))))..........))))))))................((((((...........)))))).......... (-12.24 = -12.92 + 0.68)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:27:32 2011