| Sequence ID | dm3.chr3R |
|---|---|
| Location | 25,266,142 – 25,266,276 |
| Length | 134 |
| Max. P | 0.872086 |

| Location | 25,266,142 – 25,266,249 |
|---|---|
| Length | 107 |
| Sequences | 9 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 69.07 |
| Shannon entropy | 0.59407 |
| G+C content | 0.44271 |
| Mean single sequence MFE | -18.81 |
| Consensus MFE | -10.52 |
| Energy contribution | -10.54 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.12 |
| Mean z-score | -0.92 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.808906 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
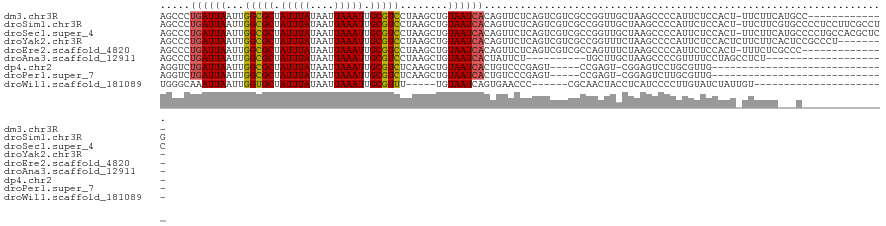
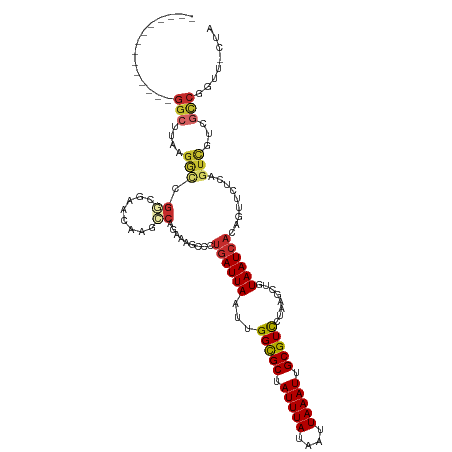
>dm3.chr3R 25266142 107 - 27905053 AGCCCUGAUUAAUUGGCGCUAUUUAUAAUUAAAUUGCGUCCUAAGCUGUAAUCACAGUUCUCAGUCGUCGCCGGUUGCUAAGCCCCAUUCUCCACU-UUCUUCAUGCC------------- .((..(((((....(((((.(((((....))))).)))))...((((((....))))))...)))))..)).((((....))))............-...........------------- ( -20.80, z-score = -1.31, R) >droSim1.chr3R 24941642 120 - 27517382 AGCCCUGAUUAAUUGGCGCUAUUUAUAAUUAAAUUGCGUCCUAAGCUGUAAUCACAGUUCUCAGUCGUCGCCGGUUGCUAAGCCCCAUUCUCCACU-UUCUUCGUGCCCCUCCUUCGCCUG .((..(((((....(((((.(((((....))))).)))))...((((((....))))))...)))))..)).((((....))))........(((.-......)))............... ( -21.30, z-score = -1.03, R) >droSec1.super_4 4124977 120 - 6179234 AGCCCUGAUUAAUUGGCGCUAUUUAUAAUUAAAUUGCGUCCUAAGCUGUAAUCACAGUUCUCAGUCGUCGCCGGUUGCUAAGCCCCAUUCUCCACU-UUCUUCAUGCCCCUGCCACGCUCC (((.((((......(((((.(((((....))))).)))))...((((((....)))))).))))..((.((.((..((..................-........)).)).)).))))).. ( -23.07, z-score = -1.20, R) >droYak2.chr3R 7612562 113 + 28832112 AGCCCUGAUUAAUUGGCGCUAUUUAUAAUUAAAUUGCGUCCUAAGCUGUAAUCACAGUUCUCAGUCGUCGCCGGUUUCUAAGCCCCAUUCUCCACUCUUCUUCACUCCGCCCU-------- .((..(((((....(((((.(((((....))))).)))))...((((((....))))))...)))))..)).((((....)))).............................-------- ( -19.70, z-score = -1.63, R) >droEre2.scaffold_4820 7727720 106 + 10470090 AGCCCUGAUUAAUUGGCGCUAUUUAUAAUUAAAUUGCGUCCUAAGCUGUAAUCACAGUUCUCAGUCGUCGCCAGUUUCUAAGCCCCAUUCUCCACU-UUUCUCGCCC-------------- .((...((..(((((((((.(((((....))))).))......((((((....))))))..........)))))))))...)).............-..........-------------- ( -18.90, z-score = -2.12, R) >droAna3.scaffold_12911 4955140 91 - 5364042 AGCCCUGAUUAAUUGGCGCUAUUUAUAAUUAAAUUGCGUCCUAAGCUGUAAUCACUAUUCU----------UGCUUGCUAAGCCCCGUUUUCCUAGCCUCU-------------------- .....((((((.(((((((.(((((....))))).))).)).))....)))))).......----------.(((.(..((((...))))..).)))....-------------------- ( -14.40, z-score = -0.36, R) >dp4.chr2 2120634 86 - 30794189 AGGUCUGAUUAAUUGGCGCUAUUUAUAAUUAAAUUGCGUCUCAAGCUGUAAUCACUGUCCCGAGU-----CCGAGU-CGGAGUCCUGCGUUG----------------------------- .((.(((((((...(((((.(((((....))))).)))))........))))))..).)).((.(-----(((...-)))).))........----------------------------- ( -20.20, z-score = -0.87, R) >droPer1.super_7 2288403 86 - 4445127 AGGUCUGAUUAAUUGGCGCUAUUUAUAAUUAAAUUGCGUCUCAAGCUGUAAUCACUGUCCCGAGU-----CCGAGU-CGGAGUCUUGCGUUG----------------------------- .((.(((((((...(((((.(((((....))))).)))))........))))))..).)).((.(-----(((...-)))).))........----------------------------- ( -20.60, z-score = -1.02, R) >droWil1.scaffold_181089 8041760 88 - 12369635 UGGGCAAAUUAAUUGGUGCUAUUUAUAAUUAAAUUGCGUUU-----UGUAAUCAGUGAACCC------CGCAACUACCUCAUCCCCUUGUAUCUAUUGU---------------------- .((((((.(((((((..........))))))).))))....-----........(((.....------))).....)).....................---------------------- ( -10.30, z-score = 1.23, R) >consensus AGCCCUGAUUAAUUGGCGCUAUUUAUAAUUAAAUUGCGUCCUAAGCUGUAAUCACAGUUCUCAGUCGUCGCCGGUUGCUAAGCCCCAUUCUCCACU_UUCUUC___C______________ .....((((((...(((((.(((((....))))).)))))........))))))................................................................... (-10.52 = -10.54 + 0.03)

| Location | 25,266,169 – 25,266,276 |
|---|---|
| Length | 107 |
| Sequences | 9 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 70.80 |
| Shannon entropy | 0.57507 |
| G+C content | 0.44649 |
| Mean single sequence MFE | -31.24 |
| Consensus MFE | -13.03 |
| Energy contribution | -13.56 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.872086 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
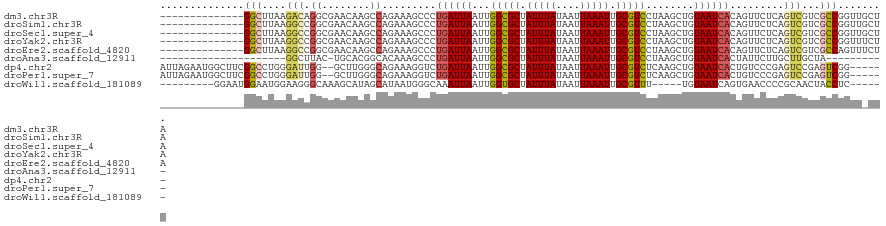
>dm3.chr3R 25266169 107 - 27905053 --------------GGCUUAAGACAGGCGAACAAGCCAGAAAGCCCUGAUUAAUUGGCGCUAUUUAUAAUUAAAUUGCGUCCUAAGCUGUAAUCACAGUUCUCAGUCGUCGCCGGUUGCUA --------------(((....(((.(((......))).((.(((..((((((.(((((((.(((((....))))).))).)).))....))))))..))).)).)))...)))........ ( -30.00, z-score = -1.47, R) >droSim1.chr3R 24941682 107 - 27517382 --------------GGCUUAAGGCCGGCGAACAAGCCAGAAAGCCCUGAUUAAUUGGCGCUAUUUAUAAUUAAAUUGCGUCCUAAGCUGUAAUCACAGUUCUCAGUCGUCGCCGGUUGCUA --------------(((....(((((((((....((......)).((((......(((((.(((((....))))).)))))...((((((....)))))).))))...)))))))))))). ( -34.10, z-score = -2.18, R) >droSec1.super_4 4125017 107 - 6179234 --------------GGCUUAAGGCCGGCGAACAAGCCAGAAAGCCCUGAUUAAUUGGCGCUAUUUAUAAUUAAAUUGCGUCCUAAGCUGUAAUCACAGUUCUCAGUCGUCGCCGGUUGCUA --------------(((....(((((((((....((......)).((((......(((((.(((((....))))).)))))...((((((....)))))).))))...)))))))))))). ( -34.10, z-score = -2.18, R) >droYak2.chr3R 7612595 107 + 28832112 --------------GGCUUAAGGCCGGCGAACAAGCCAGAAAGCCCUGAUUAAUUGGCGCUAUUUAUAAUUAAAUUGCGUCCUAAGCUGUAAUCACAGUUCUCAGUCGUCGCCGGUUUCUA --------------......((((((((((....((......)).((((......(((((.(((((....))))).)))))...((((((....)))))).))))...))))))))))... ( -33.00, z-score = -2.36, R) >droEre2.scaffold_4820 7727746 107 + 10470090 --------------GGCUUAAGGCCGGCGAACAAGCCAGAAAGCCCUGAUUAAUUGGCGCUAUUUAUAAUUAAAUUGCGUCCUAAGCUGUAAUCACAGUUCUCAGUCGUCGCCAGUUUCUA --------------(((.....)))(((((....((......)).((((......(((((.(((((....))))).)))))...((((((....)))))).))))...)))))........ ( -30.40, z-score = -1.99, R) >droAna3.scaffold_12911 4955161 89 - 5364042 ---------------------GGCUUAC-UGCACGGCACAAAGCCCUGAUUAAUUGGCGCUAUUUAUAAUUAAAUUGCGUCCUAAGCUGUAAUCACUAUUCUUGCUUGCUA---------- ---------------------((((((.-..((.(((.....))).)).......(((((.(((((....))))).))))).))))))((((.((.......)).))))..---------- ( -20.50, z-score = -1.11, R) >dp4.chr2 2120646 113 - 30794189 AUUAGAAUGGCUUCGGCCUGGGAUUGG--GCUUGGGCAGAAAGGUCUGAUUAAUUGGCGCUAUUUAUAAUUAAAUUGCGUCUCAAGCUGUAAUCACUGUCCCGAGUCCGAGUCGG------ ........(((....)))..(..((((--((((((((((...((..(((......(((((.(((((....))))).))))))))..)).......))).)))))))))))..)..------ ( -41.30, z-score = -3.15, R) >droPer1.super_7 2288415 113 - 4445127 AUUAGAAUGGCUUCGGCCUGGGAUUGG--GCUUGGGCAGAAAGGUCUGAUUAAUUGGCGCUAUUUAUAAUUAAAUUGCGUCUCAAGCUGUAAUCACUGUCCCGAGUCCGAGUCGG------ ........(((....)))..(..((((--((((((((((...((..(((......(((((.(((((....))))).))))))))..)).......))).)))))))))))..)..------ ( -41.30, z-score = -3.15, R) >droWil1.scaffold_181089 8041779 101 - 12369635 ---------GGAAUGGAAUGGAAGGGCAAAGCAUAGCAUAAUGGGCAAAUUAAUUGGUGCUAUUUAUAAUUAAAUUGCGUUU-----UGUAAUCAGUGAACCCCGCAACUACCUC------ ---------..........((.((.(((((((....(.....).((((.(((((((..........))))))).))))))))-----))).....(((.....)))..)).))..------ ( -16.50, z-score = 1.65, R) >consensus ______________GGCUUAAGGCCGGCGAACAAGCCAGAAAGCCCUGAUUAAUUGGCGCUAUUUAUAAUUAAAUUGCGUCCUAAGCUGUAAUCACAGUUCUCAGUCGUCGCCGGUU_CUA ..............(((....(((.((........)).........((((((...(((((.(((((....))))).)))))........)))))).........)))...)))........ (-13.03 = -13.56 + 0.53)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:54:31 2011