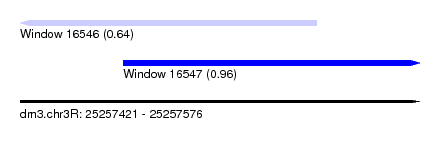
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 25,257,421 – 25,257,576 |
| Length | 155 |
| Max. P | 0.961884 |

| Location | 25,257,421 – 25,257,536 |
|---|---|
| Length | 115 |
| Sequences | 8 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 79.78 |
| Shannon entropy | 0.38625 |
| G+C content | 0.41777 |
| Mean single sequence MFE | -32.15 |
| Consensus MFE | -17.30 |
| Energy contribution | -16.32 |
| Covariance contribution | -0.98 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.643588 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 25257421 115 - 27905053 CUUUUUCCUCAUCCGGGCAAAAAGG--GUGGGAAAAAUAUGAAAAAUUGCAACAUAUUUUCCUUAGCCUCUGGCCGGAGUUAAACUUUUAUUUGUUUGCUUCAUUGCAUGGCAGAGG ......((((.(((((.((...(((--.((((.((((((((...........)))))))).)))).))).)).)))))..............(((((((......))).)))))))) ( -33.50, z-score = -1.95, R) >droSim1.chr3R 24933078 115 - 27517382 CUUUUUCCCCAUCCGGGCAAAAAGG--GUGGGAAAAAUAUGAAAAAUUGCAACAUAUUUUCCUUAGCCUCUGGCCGGAGUUAAACUUUUAUUUGUUUGCUUCAUUGCAUGGCGGAGG ....((((((((.(((((....(((--.((((.((((((((...........)))))))).)))).)))...)))(((((.((((........)))))))))..)).)))).)))). ( -33.80, z-score = -1.48, R) >droSec1.super_4 4116463 114 - 6179234 CUUUUUCCCCAUCCGGGCAAAAAGG--GUGGG-AAAAUAUGAAAAAUUGCAACAUAUUUUCCUUAGCCUCUGGCCGGAGUUAAACUUUUAUUUGUUUGCUUCAUUGCAUGGCGGAGG ....((((((((.(((((....(((--(((((-((((((((...........)))))))))))..))).)).)))(((((.((((........)))))))))..)).)))).)))). ( -36.80, z-score = -2.33, R) >droYak2.chr3R 7603564 117 + 28832112 GUUUUUCUCCAUCCGGGCAAAAAGGGUGAGAGAAGAAUAUGAAAAAUUGCAACAUAUUUUCCUUAGCCUCUGGACGGAGUUAAACUUUUAUUUGUUUGCUUCAUUGCAUGGCGGAGG ((((..((((.(((((......(((.((((.(((((.((((...........))))))))))))).)))))))).))))..)))).....(((((((((......))).)))))).. ( -33.40, z-score = -1.83, R) >droEre2.scaffold_4820 7718822 114 + 10470090 GUUUUUCCCCAUCCGGCCAAAAGG---GUGAGAAGAAUAUGAAAAAUUGCAACAUAUUUUCCUUAGCCUCUGGCCGGAGUUAAACUUUUAUUUGUUUGCUUCAUUGCAUGGCGGAGG ....(((((((((((((((..(((---.((((.((((((((...........)))))))).)))).))).))))))))..................(((......)))))).)))). ( -39.50, z-score = -3.51, R) >droAna3.scaffold_12911 4946685 98 - 5364042 ---------------GGCGGAAG----GGGUGAAAAAUAUGAAAAAUUGCAACAUAUUUUCCUUAGCCUCUGGUCUGAGUUAAACUUUUAUUUGUUUGCUUCAUUGCAUGGUCGAGG ---------------(((((.((----((((..((((((((...........)))))))).....))))))..))((((.(((((........))))).)))).......))).... ( -19.20, z-score = 0.44, R) >dp4.chr2 2111817 103 - 30794189 ----------CUCUGCGCGUGGU----AUGGGGAAAAUAUGAAAAAUUGCAACAUAUUUUCCUCGACUCCGUCUGGGGGCGAAAGUUUUAUUUGUUUGCUUCAUUGCAUGGUCGAGG ----------(((((.(((..((----.(((((((((((((...........)))))))))))))))..))).)))))((....))....((((..(((......)))....)))). ( -30.50, z-score = -1.98, R) >droPer1.super_7 2279594 103 - 4445127 ----------CUCUGCGCGUGGU----AUGGGGAAAAUAUGAAAAAUUGCAACAUAUUUUCCUCGACUCCGUCUGGGGGCGAAAGUUUUAUUUGUUUGCUUCAUUGCAUGGUCGAGG ----------(((((.(((..((----.(((((((((((((...........)))))))))))))))..))).)))))((....))....((((..(((......)))....)))). ( -30.50, z-score = -1.98, R) >consensus _UUUUUCCCCAUCCGGGCAAAAAG___GUGGGAAAAAUAUGAAAAAUUGCAACAUAUUUUCCUUAGCCUCUGGCCGGAGUUAAACUUUUAUUUGUUUGCUUCAUUGCAUGGCGGAGG .................................((((((((...........))))))))((((.(((.......(((((.((((........))))))))).......))).)))) (-17.30 = -16.32 + -0.98)

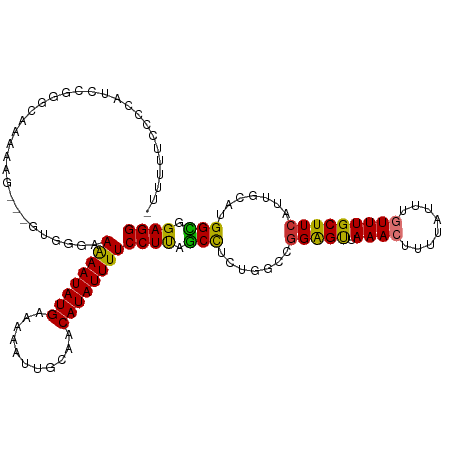
| Location | 25,257,461 – 25,257,576 |
|---|---|
| Length | 115 |
| Sequences | 7 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 70.85 |
| Shannon entropy | 0.52783 |
| G+C content | 0.47757 |
| Mean single sequence MFE | -30.20 |
| Consensus MFE | -19.44 |
| Energy contribution | -19.20 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.961884 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 25257461 115 + 27905053 -CCGGCCAGAGGCUAAGGAAAAUAUGUUGCAAUUUUUCAUAUUUUUCCCAC--CCUUUUUGCCCGGAUGAGGAAAAAGCAUCCAGCUUCUUGCCCGGCAUCUCGAAAUCCGCUACAGA -(((((.(((((((..((((((((((...........))))))))))....--...........(((((.........)))))))))))).).))))..................... ( -29.30, z-score = -0.56, R) >droSim1.chr3R 24933118 115 + 27517382 -CCGGCCAGAGGCUAAGGAAAAUAUGUUGCAAUUUUUCAUAUUUUUCCCAC--CCUUUUUGCCCGGAUGGGGAAAAAGCAUCCAGCUUCUUGCCCGGCAUCUAGAAAUCCGCUGCAGA -..(((.(((((((..((((((((((...........))))))))))....--.((((((.(((....))).)))))).....))))))).)))((((............)))).... ( -34.20, z-score = -0.79, R) >droSec1.super_4 4116503 114 + 6179234 -CCGGCCAGAGGCUAAGGAAAAUAUGUUGCAAUUUUUCAUAUUUU-CCCAC--CCUUUUUGCCCGGAUGGGGAAAAAGCAUCCAGCUUCUUGCCCGGCAUCUCGAAAUCCGCUGCAGA -..(((.(((((((..((((((((((...........))))))))-))...--.((((((.(((....))).)))))).....))))))).)))((((............)))).... ( -37.10, z-score = -1.61, R) >droYak2.chr3R 7603604 117 - 28832112 -CCGUCCAGAGGCUAAGGAAAAUAUGUUGCAAUUUUUCAUAUUCUUCUCUCACCCUUUUUGCCCGGAUGGAGAAAAACCAUUCAGCUUCUUGCCCGGCAACUCAAAAUCCGCUGCAGA -(((..((((((((.((((.((((((...........)))))).))))................((((((.......)))))))))))).))..)))..................... ( -24.00, z-score = 0.38, R) >droEre2.scaffold_4820 7718862 114 - 10470090 -CCGGCCAGAGGCUAAGGAAAAUAUGUUGCAAUUUUUCAUAUUCUUCUCAC---CCUUUUGGCCGGAUGGGGAAAAACCAUUCAGCUUCUUGCCCGGCAACUCAAAAUCCGCUGCAGA -(((((((((((...((((.((((((...........)))))).))))...---.)))))))))))((((.......)))).((((.........(....).........)))).... ( -31.97, z-score = -0.64, R) >dp4.chr2 2111857 88 + 30794189 CCCAGACGGAGUC-GAGGAAAAUAUGUUGCAAUUUUUCAUAUUUUCCCCAU----------ACCACGCGCAGAGCAGCUCUGCUGAAAUCCGCUCCAGA------------------- .......(((((.-((((((((((((...........))))))))))....----------.......((((((...)))))).....)).)))))...------------------- ( -27.40, z-score = -3.54, R) >droPer1.super_7 2279634 88 + 4445127 CCCAGACGGAGUC-GAGGAAAAUAUGUUGCAAUUUUUCAUAUUUUCCCCAU----------ACCACGCGCAGAGCAGCUCUGCUGAAAUCCGCUCCAGA------------------- .......(((((.-((((((((((((...........))))))))))....----------.......((((((...)))))).....)).)))))...------------------- ( -27.40, z-score = -3.54, R) >consensus _CCGGCCAGAGGCUAAGGAAAAUAUGUUGCAAUUUUUCAUAUUUUUCCCAC__CCUUUUUGCCCGGAUGGGGAAAAACCAUCCAGCUUCUUGCCCGGCAUCUCGAAAUCCGCUGCAGA ...(((.(((((((..((((((((((...........)))))))))).................(((((.........)))))))))))).)))........................ (-19.44 = -19.20 + -0.24)
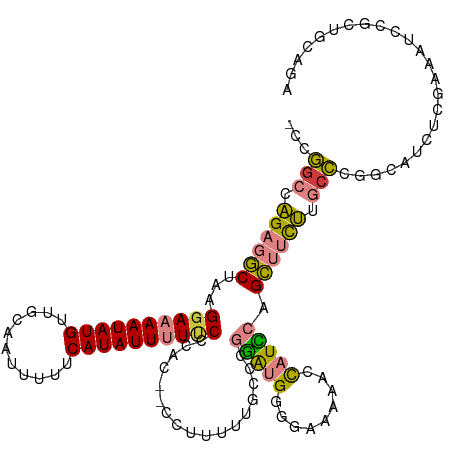
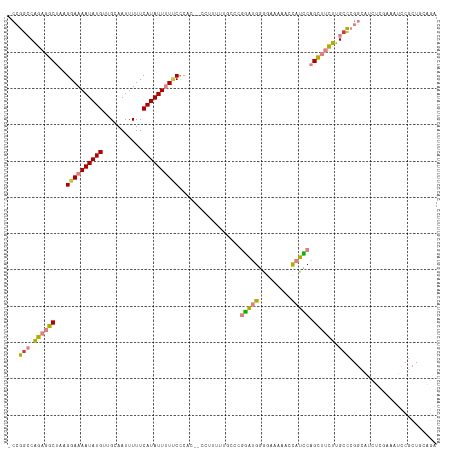
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:54:29 2011