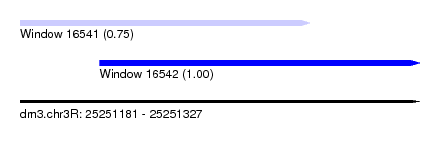
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 25,251,181 – 25,251,327 |
| Length | 146 |
| Max. P | 0.998335 |

| Location | 25,251,181 – 25,251,287 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 71.25 |
| Shannon entropy | 0.49469 |
| G+C content | 0.46098 |
| Mean single sequence MFE | -26.47 |
| Consensus MFE | -15.56 |
| Energy contribution | -16.73 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.749954 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
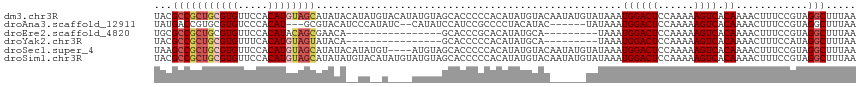
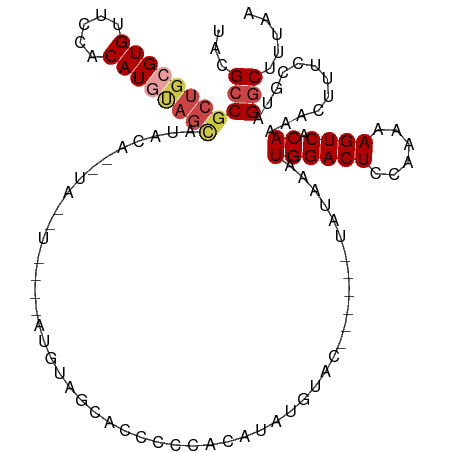
>dm3.chr3R 25251181 106 + 27905053 -------GUGUGGCUGCCACAGACAAUGAUUAAUCGUACGCCGCUGCGUGUUCCACAUGUAGCAUAUACAUAUGUACAUAUGUAGCACCCCCACAUAUGUACAAUAUGUAUAA -------.(((((...)))))....((((....)))).....((((((((.....)))))))).(((((((((((((((((((.(.....).)))))))))).))))))))). ( -37.10, z-score = -3.13, R) >droAna3.scaffold_12911 4940110 102 + 5364042 UCAGUUCCUGCUGCUGCCAGAGACAAUGAUUAAUCGUAUGACCGUGCGUGUCCCACAUGC-GUACAUCC-CAUAUCCAUAU---CCAUCCGCCCCUACAUACUAUAA------ ...((..((((....).)))..))...........(((((...((((((((.....))))-))))....-.(((....)))---.............))))).....------ ( -15.70, z-score = -0.12, R) >droEre2.scaffold_4820 7712621 81 - 10470090 -------GUGCGGCUGCCACAGACAAUGAUUAAUCGUGCGCCGCUGCGUGUUCCACAUACAGCGAACA----------------GCACCCGCACAUAUGCAUAA--------- -------((((((.(((....(...((((....)))).)..(((((..((.....))..)))))....----------------))).))))))..........--------- ( -26.80, z-score = -1.40, R) >droYak2.chr3R 7597305 81 - 28832112 -------GUGUGGCUGCCACAGACAAUGAUUAAUCGUACGCCGCUGCGUGUUUCACAUGUAGUAUACA----------------GCACCCCCACAUAUGCAUAA--------- -------((((((.(((........((((....)))).....((((((((.....)))))))).....----------------)))...))))))........--------- ( -20.00, z-score = -0.11, R) >droSec1.super_4 4110298 102 + 6179234 -------GUGUGACUGCCACAGACAAUGAUUAAUCGUAAGCCGCUGCGUGUUCCACAUGUAGCAUAUACAUAUGU----AUGUAGCACCCCCACAUAUGUACAAUAUGUAUAA -------.((((.....))))....((((....)))).....((((((((.....)))))))).(((((((((((----(((((...........))))))).))))))))). ( -27.60, z-score = -1.49, R) >droSim1.chr3R 24926878 106 + 27517382 -------GUGUGGCUGCCACAGACAAUGAUUAAUCGUACGCCGCUGCGUGUUCCACAUGUAGCAUAUAUGUACAUAUGUAUGUAGCACCCCCACAUAUGUACAAUAUGUAUAA -------((((((.(((.(((.(((..((....))(((((..((((((((.....)))))))).....)))))...))).))).)))...)))))).(((((.....))))). ( -31.60, z-score = -1.01, R) >consensus _______GUGUGGCUGCCACAGACAAUGAUUAAUCGUACGCCGCUGCGUGUUCCACAUGUAGCAUAUAC_UAU_U____AU___GCACCCCCACAUAUGUACAAUAU______ .......(((.((.(((........((((....)))).....((((((((.....)))))))).....................))).)).)))................... (-15.56 = -16.73 + 1.17)
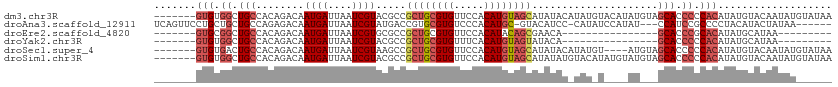
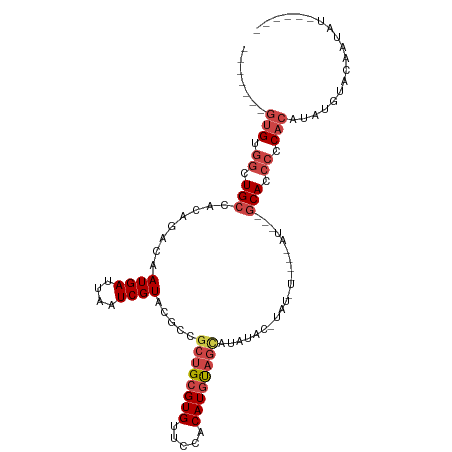
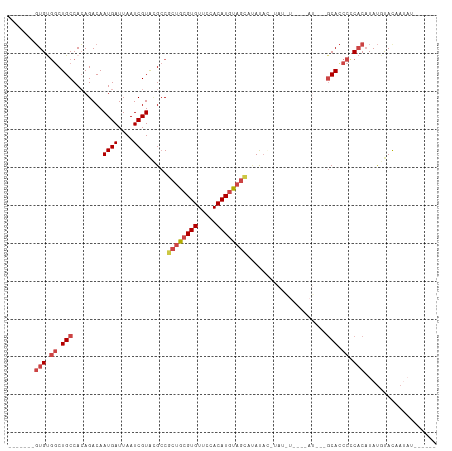
| Location | 25,251,210 – 25,251,327 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 76.76 |
| Shannon entropy | 0.43390 |
| G+C content | 0.42021 |
| Mean single sequence MFE | -27.85 |
| Consensus MFE | -16.10 |
| Energy contribution | -17.05 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.19 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.33 |
| SVM RNA-class probability | 0.998335 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 25251210 117 + 27905053 UACGCCGCUGCGUGUUCCACAUGUAGCAUAUACAUAUGUACAUAUGUAGCACCCCCACAUAUGUACAAUAUGUAUAAAUGGACUCCAAAAAGUCACAAAACUUUCCGUAGGCUUUAA ...(((((((((((.....)))))))).(((((((((((((((((((.(.....).)))))))))).)))))))))..((((((......)))).))............)))..... ( -41.00, z-score = -6.44, R) >droAna3.scaffold_12911 4940146 106 + 5364042 UAUGACCGUGCGUGUCCCACAU---GCGUACAUCCCAUAUC--CAUAUCCAUCCGCCCCUACAUAC------UAUAAAUGGACUCCAAAAAGUCACAAAACUUUCCGUAGGCUUUAA .......((((((((.....))---))))))..........--..............(((((....------........((((......))))............)))))...... ( -16.95, z-score = -1.38, R) >droEre2.scaffold_4820 7712650 92 - 10470090 UGCGCCGCUGCGUGUUCCACAUACAGCGAACA----------------GCACCCGCACAUAUGCA---------UAAAUGGACUCCAAAAAGUCACAAAACUUUCCGUAGGCUUUAA ...(((((((..((.....))..)))).....----------------((....(((....))).---------....((((((......)))).)).........)).)))..... ( -20.10, z-score = -0.64, R) >droYak2.chr3R 7597334 92 - 28832112 UACGCCGCUGCGUGUUUCACAUGUAGUAUACA----------------GCACCCCCACAUAUGCA---------UAAAUGGACUCCAAAAAGUCACAAAACUUUCCAUAGGCUUUAA ...(((((((((((.....)))))))).....----------------(((..........))).---------...(((((.......((((......))))))))).)))..... ( -19.71, z-score = -1.95, R) >droSec1.super_4 4110327 113 + 6179234 UAAGCCGCUGCGUGUUCCACAUGUAGCAUAUACAUAUGU----AUGUAGCACCCCCACAUAUGUACAAUAUGUAUAAAUGGACUCCAAAAAGUCACAAAACUUUCCGUAGGCUUUAA .(((((((((((((.....)))))))).(((((((((((----(((((...........))))))).)))))))))..((((((......)))).))............)))))... ( -34.50, z-score = -4.75, R) >droSim1.chr3R 24926907 117 + 27517382 UACGCCGCUGCGUGUUCCACAUGUAGCAUAUAUGUACAUAUGUAUGUAGCACCCCCACAUAUGUACAAUAUGUAUAAAUGGACUCCAAAAAGUCACAAAACUUUCCGUAGGCUUUAA ...(((((((((((.....))))))))((((((((((((((((.............)))))))))).)))))).....((((((......)))).))............)))..... ( -34.82, z-score = -3.98, R) >consensus UACGCCGCUGCGUGUUCCACAUGUAGCAUACA__UA__U____AUGUAGCACCCCCACAUAUGUAC______UAUAAAUGGACUCCAAAAAGUCACAAAACUUUCCGUAGGCUUUAA ...(((((((((((.....))))))))...................................................((((((......)))).))............)))..... (-16.10 = -17.05 + 0.95)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:54:25 2011