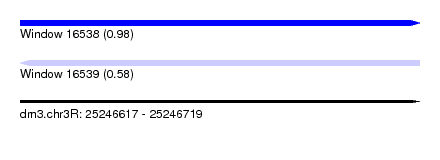
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 25,246,617 – 25,246,719 |
| Length | 102 |
| Max. P | 0.976016 |

| Location | 25,246,617 – 25,246,719 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 84.49 |
| Shannon entropy | 0.26231 |
| G+C content | 0.48471 |
| Mean single sequence MFE | -33.86 |
| Consensus MFE | -21.60 |
| Energy contribution | -22.12 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.95 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.976016 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 25246617 102 + 27905053 CUCCAUUUAAAUAGAAAAGAAAGCACAGGCACGCGUAUAAAGCCAUCUGUGCAUCGAUGAGCAUGUGUGUGUG------------CGAAGGGUGUUCCCCCGGUUUAAGUCGAG (((.((((((((..........(((((.((((((.......)).....((((........)))))))).))))------------)...(((......))).)))))))).))) ( -30.90, z-score = -2.51, R) >droSec1.super_4 4105695 101 + 6179234 CUCCAUUUAAAUAGAAAAGAAAGCACAGGCACGCGUAUAAAGCCAUCUGUGCAUCGAUGGGCAUGUGUGUGUG------------CGAAGGGGGUUCCCCGG-UUUAAGUCGAG (((.((((((((..........(((((.(((((.((......(((((........))))))).))))).))))------------)...((((...)))).)-))))))).))) ( -34.40, z-score = -2.73, R) >droYak2.chr3R 7592699 113 - 28832112 CUCCAUUUAAAUAGAAAAGAAAGCACAGGCGCGCGUAUAAAGCCAUCUGUGCAUCGAUGAGCAUGUGUGUGUGUGUAUGUGUGUGCGAAGGGGGUUCCCCCG-UUUAAGUCGAG (((.((((((((......(...(((((.(((((((((((..((((((........)))).))...))))))))))).)))))...)...((((....)))))-))))))).))) ( -40.60, z-score = -4.12, R) >droEre2.scaffold_4820 7702825 105 - 10470090 CUCCAUUUAAAUAGAAAAGAAAGCACAGGCACGCGUAUAAAGCCAUCUGUGCAUCGAUGAGCAUGUGUGUGUGUGUA--------CGAAGGGGUUUCCCCCG-UUUAAGUCGAG (((.((((((((..........(((((.((((((((.....((((((........)))).)))))))))).))))).--------....((((....)))))-))))))).))) ( -37.00, z-score = -4.55, R) >droAna3.scaffold_12911 4935340 95 + 5364042 CUCCAUUUAAAUAGAAAAGAAAACACUGGCGGGCGCAUAAAGCCAUCUGUGCAUCAGUGUGUGGGUGGGUGUG----------------CGGGGUUCC---GUGUUAAGUCAAG ..................((.(((((.((....((((((...(((((((..((....))..))))))).))))----------------)).....))---)))))...))... ( -26.40, z-score = -0.83, R) >consensus CUCCAUUUAAAUAGAAAAGAAAGCACAGGCACGCGUAUAAAGCCAUCUGUGCAUCGAUGAGCAUGUGUGUGUG____________CGAAGGGGGUUCCCCCG_UUUAAGUCGAG (((.(((((((...........(((((.(((.((.(((...(((....).))....))).)).))).))))).................((((....))))..))))))).))) (-21.60 = -22.12 + 0.52)

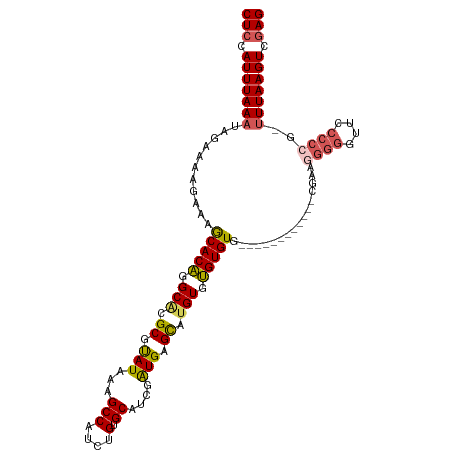
| Location | 25,246,617 – 25,246,719 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 84.49 |
| Shannon entropy | 0.26231 |
| G+C content | 0.48471 |
| Mean single sequence MFE | -23.71 |
| Consensus MFE | -16.40 |
| Energy contribution | -17.04 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.577743 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 25246617 102 - 27905053 CUCGACUUAAACCGGGGGAACACCCUUCG------------CACACACACAUGCUCAUCGAUGCACAGAUGGCUUUAUACGCGUGCCUGUGCUUUCUUUUCUAUUUAAAUGGAG (((.(.(((((..((((.....))))..(------------((((..(((.....((((........))))((.......)))))..)))))...........))))).).))) ( -24.50, z-score = -1.72, R) >droSec1.super_4 4105695 101 - 6179234 CUCGACUUAAA-CCGGGGAACCCCCUUCG------------CACACACACAUGCCCAUCGAUGCACAGAUGGCUUUAUACGCGUGCCUGUGCUUUCUUUUCUAUUUAAAUGGAG (((.(.(((((-..((((...))))...(------------((((....((((((((((........)))))........)))))..)))))...........))))).).))) ( -25.90, z-score = -2.23, R) >droYak2.chr3R 7592699 113 + 28832112 CUCGACUUAAA-CGGGGGAACCCCCUUCGCACACACAUACACACACACACAUGCUCAUCGAUGCACAGAUGGCUUUAUACGCGCGCCUGUGCUUUCUUUUCUAUUUAAAUGGAG .((((......-.((((....))))...(((....................)))...)))).((((((.((((.......)).)).)))))).......(((((....))))). ( -25.25, z-score = -2.12, R) >droEre2.scaffold_4820 7702825 105 + 10470090 CUCGACUUAAA-CGGGGGAAACCCCUUCG--------UACACACACACACAUGCUCAUCGAUGCACAGAUGGCUUUAUACGCGUGCCUGUGCUUUCUUUUCUAUUUAAAUGGAG (((.(.(((((-.((((....))))...(--------((((..(((.(....((.((((........)))))).......).)))..)))))...........))))).).))) ( -28.80, z-score = -3.17, R) >droAna3.scaffold_12911 4935340 95 - 5364042 CUUGACUUAACAC---GGAACCCCG----------------CACACCCACCCACACACUGAUGCACAGAUGGCUUUAUGCGCCCGCCAGUGUUUUCUUUUCUAUUUAAAUGGAG ...((...(((((---((.....((----------------((.......(((....(((.....))).))).....))))....)).))))).))...(((((....))))). ( -14.10, z-score = 0.32, R) >consensus CUCGACUUAAA_CGGGGGAACCCCCUUCG____________CACACACACAUGCUCAUCGAUGCACAGAUGGCUUUAUACGCGUGCCUGUGCUUUCUUUUCUAUUUAAAUGGAG (((.(.((((...((((....))))..................................((.((((((..(((.......)).)..))))))..))........)))).).))) (-16.40 = -17.04 + 0.64)

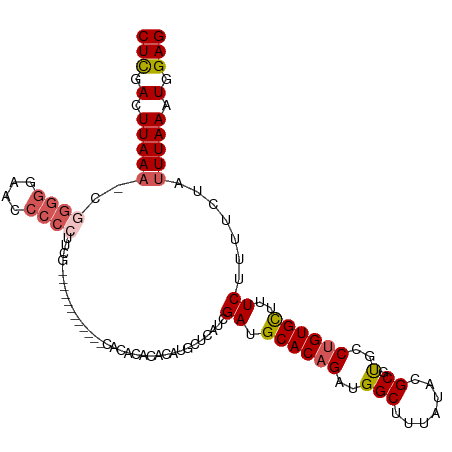
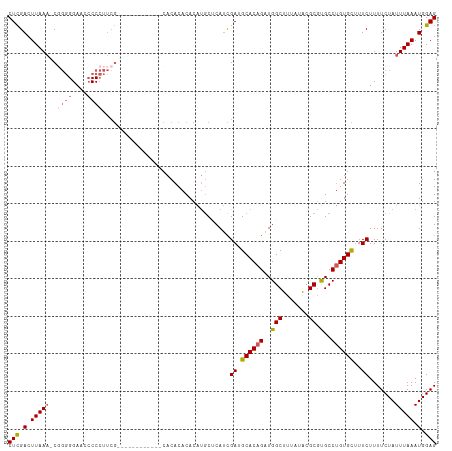
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:54:22 2011