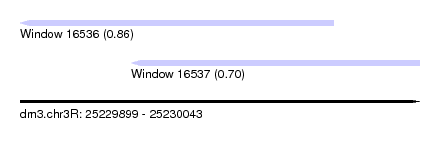
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 25,229,899 – 25,230,043 |
| Length | 144 |
| Max. P | 0.857714 |

| Location | 25,229,899 – 25,230,012 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.82 |
| Shannon entropy | 0.24488 |
| G+C content | 0.39757 |
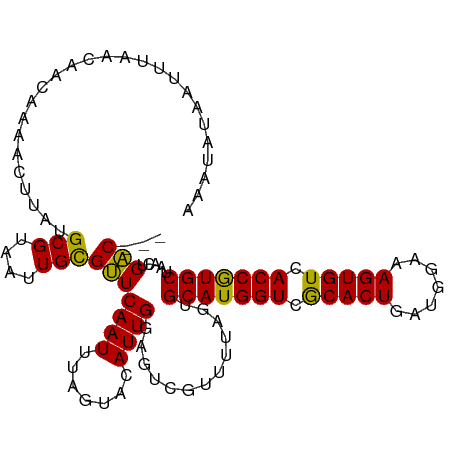
| Mean single sequence MFE | -32.31 |
| Consensus MFE | -22.39 |
| Energy contribution | -22.40 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.857714 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 25229899 113 - 27905053 AUUGUGAUCAAUGUAUUACAUUGG-------CUACUGCAUGGUCACACUGGUGGAAAGUGUGACCAUGCUAACUGUCAAACGCGACGUUUUUGUACCUAAGUAACUUUUUCUUUGCGUGC ...(((..(((((.....))))).-------.))).(((((((((((((.......)))))))))))))..........((((((.(......(((....))).......).)))))).. ( -38.52, z-score = -3.82, R) >droSec1.super_4 4090105 112 - 6179234 AUUGCGUUCAAUUUAGUACAUUGGAGUCGUUUUAGUGCAUGGUCGCACUGAUGGAAAGUGUCACCGUGCUAACUGAC--------UGUUUUUUUACUUAAGGAACUUUUUCUUUACGUGU .....((((.....((((....(.(((((((.....(((((((.(((((.......))))).))))))).))).)))--------).).....))))....))))............... ( -29.20, z-score = -1.62, R) >droSim1.chr3R 24904664 112 - 27517382 AUUGCGUUCAAUUUAGUACAUUGGAGUCGUUUUAGUGCAUGGUCGCACUGAUGGAAAGUGUCACCGUGCUAACUGAC--------UGUUUUUUUACUUAAGGAACUUUUUCUUUGCGUGU .....((((.....((((....(.(((((((.....(((((((.(((((.......))))).))))))).))).)))--------).).....))))....))))............... ( -29.20, z-score = -1.36, R) >consensus AUUGCGUUCAAUUUAGUACAUUGGAGUCGUUUUAGUGCAUGGUCGCACUGAUGGAAAGUGUCACCGUGCUAACUGAC________UGUUUUUUUACUUAAGGAACUUUUUCUUUGCGUGU ......((((((.......))))))...........(((((((.(((((.......))))).)))))))........................(((.(((((((....))))))).))). (-22.39 = -22.40 + 0.01)

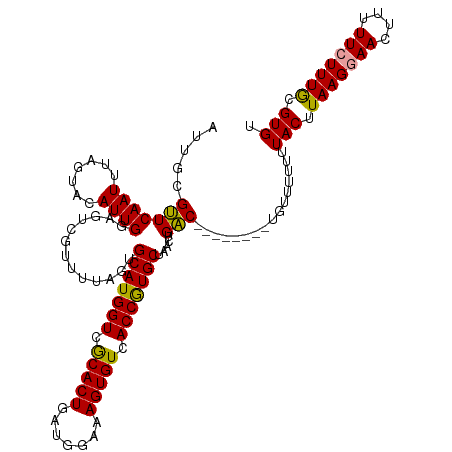
| Location | 25,229,939 – 25,230,043 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 82.12 |
| Shannon entropy | 0.24592 |
| G+C content | 0.37500 |
| Mean single sequence MFE | -27.76 |
| Consensus MFE | -20.53 |
| Energy contribution | -20.20 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.701702 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 25229939 104 - 27905053 AAAUAUACUUUCAUUACCAAACUUCUGCGUUAUUGUGAUCAAUGUAUUACAUUGG-------CUACUGCAUGGUCACACUGGUGGAAAGUGUGACCAUGCUAACUGUCAAA ............................((((..(((..(((((.....))))).-------.))).(((((((((((((.......)))))))))))))))))....... ( -33.20, z-score = -4.19, R) >droSec1.super_4 4090140 108 - 6179234 AAAUUUAAUUUAACAACAAAACUUAUGCGUAAUUGCGUUCAAUUUAGUACAUUGGAGUCGUUUUAGUGCAUGGUCGCACUGAUGGAAAGUGUCACCGUGCUAACUGAC--- ...........(((.((........((((....))))((((((.......)))))))).)))((((((((((((.(((((.......))))).)))))))..))))).--- ( -25.00, z-score = -0.79, R) >droSim1.chr3R 24904699 108 - 27517382 AAAUGUAAUUUAACAACAAAACUUAUGCGUAAUUGCGUUCAAUUUAGUACAUUGGAGUCGUUUUAGUGCAUGGUCGCACUGAUGGAAAGUGUCACCGUGCUAACUGAC--- .((((((.................(((((....))))).........)))))).........((((((((((((.(((((.......))))).)))))))..))))).--- ( -25.07, z-score = -0.46, R) >consensus AAAUAUAAUUUAACAACAAAACUUAUGCGUAAUUGCGUUCAAUUUAGUACAUUGGAGUCGUUUUAGUGCAUGGUCGCACUGAUGGAAAGUGUCACCGUGCUAACUGAC___ ..........................(((....))).((((((.......))))))...........(((((((.(((((.......))))).)))))))........... (-20.53 = -20.20 + -0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:54:21 2011