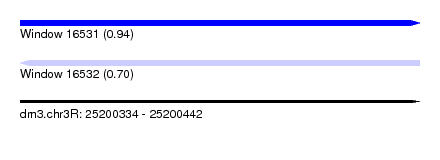
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 25,200,334 – 25,200,442 |
| Length | 108 |
| Max. P | 0.943129 |

| Location | 25,200,334 – 25,200,442 |
|---|---|
| Length | 108 |
| Sequences | 12 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 73.97 |
| Shannon entropy | 0.53462 |
| G+C content | 0.38872 |
| Mean single sequence MFE | -24.98 |
| Consensus MFE | -15.58 |
| Energy contribution | -15.60 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.943129 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3R 25200334 108 + 27905053 UUUGCGAUAAAAAAUGAUUGCACAUGCGAGCGUUUU----GGCUCGAAAUUGCAGCAUUUGCGGCAUGGAAA-UAUAUUUAGAUGUAUAUAUGCAAGGAGAUCUACUAUAGAU ....((.....(((((.(((((....(((((.....----.)))))....)))))))))).))(((((...(-(((((....)))))).)))))......(((((...))))) ( -25.60, z-score = -0.78, R) >droSim1.chr3R 24875229 108 + 27517382 UUUGCGAUAAAAAAUGAUUGCACAUGCGAGCGUUUU----GGCUCGAAAUUGCAGCAUUUGCGGCAUGGAAA-UAUAUUUAGAUGUAUAUAUGCGAGGAGAUCUACUAUAGAU ....((.....(((((.(((((....(((((.....----.)))))....)))))))))).))(((((...(-(((((....)))))).)))))......(((((...))))) ( -25.60, z-score = -0.67, R) >droSec1.super_4 4061347 108 + 6179234 UUUGCGAUAAAAAAUGAUUGCACAUGCGAGCGUUUU----GGCUCGAAAUUGCAGCAUUUGCGGCAUGGAAA-UAUAUUUAGAUGUAUAUAUGCAAGGAGAUCUACUAUAGAU ....((.....(((((.(((((....(((((.....----.)))))....)))))))))).))(((((...(-(((((....)))))).)))))......(((((...))))) ( -25.60, z-score = -0.78, R) >droYak2.chr3R 7546043 106 - 28832112 UUUGCGAUAAAAAAUGAUUGCACAUGCGAGCGUUUU----GGCUCGAAAUUGCAGCAUUUGCGGCAUGGAAA-UAUAUUUAGAUGUAUAUGGAC--GGAGAUCUAGUAUAGAU ..((((((........))))))(((((((((.....----.)))).....(((((...))))))))))....-((((((.((((.(........--..).))))))))))... ( -23.90, z-score = -0.43, R) >droEre2.scaffold_4820 7649322 106 - 10470090 UUUGCGAUAAAAAAUGAUUGCACAUGCGAGCGUUUU----GGCUCGAAAUUGCAGCAUUUGCGGCAUGGAAA-UAUAUUUAGAUAUAUAUGGAC--GGAGAUCUACUAUAGAU ..((((((........))))))(((((((((.....----.)))).....(((((...))))))))))...(-(((((......))))))....--....(((((...))))) ( -22.40, z-score = -0.40, R) >droAna3.scaffold_12911 4892088 97 + 5364042 UUUGCGAUAAAAAAUGAUUGCACAUGCGAGCGUUUU----GGCUCGAAAUUGCAGCAUUUGCGGCAUGGAAAAUAUAUUUCGCAGGCCAGA-----AGAACUCAAC------- .(((.(.....(((((.(((((....(((((.....----.)))))....))))))))))..(((.((((((.....)))).)).)))...-----....).))).------- ( -25.30, z-score = -0.82, R) >dp4.chr2 2057008 97 + 30794189 UUUGCGAUAAAAAAUGAUUGCACAUGUGAGCGUUCCG---GGCUCAAAAUUGCAGCAUUUGCGGCAUGGAAA--AUAUUUCACAGGCCAAAGGAAACUCAAC----------- ..((((((........))))))....((((..((((.---((((......(((.((....)).)))(((((.--...)))))..))))...)))).))))..----------- ( -25.20, z-score = -1.29, R) >droPer1.super_7 2224694 97 + 4445127 UUUGCGAUAAAAAAUGAUUGCACAUGUGAGCGUUCCG---GGCUCAAAAUUGCAGCAUUUGCGGCAUGGAAA--AUAUUUCACAGGCCAAAGGAAACUCAAC----------- ..((((((........))))))....((((..((((.---((((......(((.((....)).)))(((((.--...)))))..))))...)))).))))..----------- ( -25.20, z-score = -1.29, R) >droWil1.scaffold_181089 7975193 106 + 12369635 UUUGCGAUAAAAAAUGAUUGCACAUGUGAGUGUUU-----GGCUGAAAAUUGCAACAUUUGCGGCAUGAAAA-UAUAUUUCACGAGGCAGAAAGAAGAAGACGAAACUCAAC- ..((((((........))))))....(((((.(((-----(((........))....(((((..(.(((((.-....))))).)..)))))..........)))))))))..- ( -21.30, z-score = -0.82, R) >droVir3.scaffold_12822 2543622 97 + 4096053 UUUGCGAUAAAAAAAGAUUGCACAUGCGACCGUUUUG----GCACAAAAUUGCAACAUUUGCGGCAUGAAAA-UGCCGC--ACAAGCUAAAAA---GAAAACUCAAC------ ..((((((........))))))..........(((((----((.((....)).......((((((((....)-))))))--)...))))))).---...........------ ( -24.50, z-score = -2.20, R) >droMoj3.scaffold_6540 26639436 96 + 34148556 UUUGCGAUAAAAAAUGAUUGCACAUGCGAGUGUUUUG----CCUCGAAAUUGCAACAUUUGCGGCAUGAAAA-UGCCGC--ACAAGC-CAAAA---GAAAACCCAAC------ ...((......(((((.(((((....((((.(.....----)))))....))))))))))(((((((....)-))))))--....))-.....---...........------ ( -29.70, z-score = -3.67, R) >droGri2.scaffold_14624 3451565 102 + 4233967 UUUGCGAUAAAAAAUGAUUACACAUAUGAGCCGCUUGAGCCGCUCAAAAUUGCAGCAUUUGCGGCAUGAAAA-UGCCACCGACUAGA-AAAAA---AAAAACACAAA------ ..((((((.....(((......))).(((((.((....)).)))))..))))))((....))(((((....)-))))..........-.....---...........------ ( -25.50, z-score = -3.43, R) >consensus UUUGCGAUAAAAAAUGAUUGCACAUGCGAGCGUUUU____GGCUCGAAAUUGCAGCAUUUGCGGCAUGGAAA_UAUAUUUAACUGGAUAAAAAC__GGAAACCUACU______ ..((((((........))))))(((((((((..........)))).....(((((...))))))))))............................................. (-15.58 = -15.60 + 0.02)

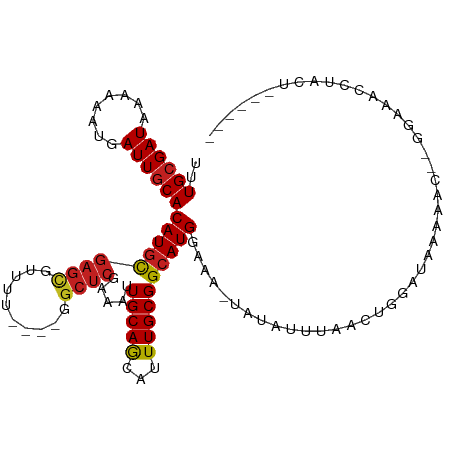
| Location | 25,200,334 – 25,200,442 |
|---|---|
| Length | 108 |
| Sequences | 12 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 73.97 |
| Shannon entropy | 0.53462 |
| G+C content | 0.38872 |
| Mean single sequence MFE | -21.41 |
| Consensus MFE | -11.86 |
| Energy contribution | -11.55 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.704249 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3R 25200334 108 - 27905053 AUCUAUAGUAGAUCUCCUUGCAUAUAUACAUCUAAAUAUA-UUUCCAUGCCGCAAAUGCUGCAAUUUCGAGCC----AAAACGCUCGCAUGUGCAAUCAUUUUUUAUCGCAAA .......(((((.....((((((((...............-......(((.((....)).)))....(((((.----.....))))).))))))))......)))))...... ( -20.20, z-score = -1.49, R) >droSim1.chr3R 24875229 108 - 27517382 AUCUAUAGUAGAUCUCCUCGCAUAUAUACAUCUAAAUAUA-UUUCCAUGCCGCAAAUGCUGCAAUUUCGAGCC----AAAACGCUCGCAUGUGCAAUCAUUUUUUAUCGCAAA (((((...)))))......((((((...............-......(((.((....)).)))....(((((.----.....))))).))))))................... ( -18.80, z-score = -1.13, R) >droSec1.super_4 4061347 108 - 6179234 AUCUAUAGUAGAUCUCCUUGCAUAUAUACAUCUAAAUAUA-UUUCCAUGCCGCAAAUGCUGCAAUUUCGAGCC----AAAACGCUCGCAUGUGCAAUCAUUUUUUAUCGCAAA .......(((((.....((((((((...............-......(((.((....)).)))....(((((.----.....))))).))))))))......)))))...... ( -20.20, z-score = -1.49, R) >droYak2.chr3R 7546043 106 + 28832112 AUCUAUACUAGAUCUCC--GUCCAUAUACAUCUAAAUAUA-UUUCCAUGCCGCAAAUGCUGCAAUUUCGAGCC----AAAACGCUCGCAUGUGCAAUCAUUUUUUAUCGCAAA ........(((((....--..........)))))......-......(((.((....))((((....(((((.----.....)))))....)))).............))).. ( -16.14, z-score = -0.97, R) >droEre2.scaffold_4820 7649322 106 + 10470090 AUCUAUAGUAGAUCUCC--GUCCAUAUAUAUCUAAAUAUA-UUUCCAUGCCGCAAAUGCUGCAAUUUCGAGCC----AAAACGCUCGCAUGUGCAAUCAUUUUUUAUCGCAAA (((((...)))))....--....((((((......)))))-).....(((.((....))((((....(((((.----.....)))))....)))).............))).. ( -16.80, z-score = -0.80, R) >droAna3.scaffold_12911 4892088 97 - 5364042 -------GUUGAGUUCU-----UCUGGCCUGCGAAAUAUAUUUUCCAUGCCGCAAAUGCUGCAAUUUCGAGCC----AAAACGCUCGCAUGUGCAAUCAUUUUUUAUCGCAAA -------..........-----...(((.((.((((.....)))))).)))(((((((.((((....(((((.----.....)))))....))))..)))))......))... ( -22.60, z-score = -0.69, R) >dp4.chr2 2057008 97 - 30794189 -----------GUUGAGUUUCCUUUGGCCUGUGAAAUAU--UUUCCAUGCCGCAAAUGCUGCAAUUUUGAGCC---CGGAACGCUCACAUGUGCAAUCAUUUUUUAUCGCAAA -----------..(((((((((...(((.((.(((....--.))))).)))(((.....)))...........---.)))).)))))....(((..............))).. ( -19.54, z-score = 0.39, R) >droPer1.super_7 2224694 97 - 4445127 -----------GUUGAGUUUCCUUUGGCCUGUGAAAUAU--UUUCCAUGCCGCAAAUGCUGCAAUUUUGAGCC---CGGAACGCUCACAUGUGCAAUCAUUUUUUAUCGCAAA -----------..(((((((((...(((.((.(((....--.))))).)))(((.....)))...........---.)))).)))))....(((..............))).. ( -19.54, z-score = 0.39, R) >droWil1.scaffold_181089 7975193 106 - 12369635 -GUUGAGUUUCGUCUUCUUCUUUCUGCCUCGUGAAAUAUA-UUUUCAUGCCGCAAAUGUUGCAAUUUUCAGCC-----AAACACUCACAUGUGCAAUCAUUUUUUAUCGCAAA -..(((((................(((..(((((((....-.)))))))..)))...(((((........)).-----.))))))))....(((..............))).. ( -14.84, z-score = 0.02, R) >droVir3.scaffold_12822 2543622 97 - 4096053 ------GUUGAGUUUUC---UUUUUAGCUUGU--GCGGCA-UUUUCAUGCCGCAAAUGUUGCAAUUUUGUGC----CAAAACGGUCGCAUGUGCAAUCUUUUUUUAUCGCAAA ------((((((.....---..))))))((((--((((((-(....)))))))(((.((((((....(((((----(.....)).))))..)))))).))).......)))). ( -28.20, z-score = -2.19, R) >droMoj3.scaffold_6540 26639436 96 - 34148556 ------GUUGGGUUUUC---UUUUG-GCUUGU--GCGGCA-UUUUCAUGCCGCAAAUGUUGCAAUUUCGAGG----CAAAACACUCGCAUGUGCAAUCAUUUUUUAUCGCAAA ------((..((.....---.))..-))((((--((((((-(....)))))))((((((((((....((((.----.......))))....))))).)))))......)))). ( -30.10, z-score = -2.40, R) >droGri2.scaffold_14624 3451565 102 - 4233967 ------UUUGUGUUUUU---UUUUU-UCUAGUCGGUGGCA-UUUUCAUGCCGCAAAUGCUGCAAUUUUGAGCGGCUCAAGCGGCUCAUAUGUGUAAUCAUUUUUUAUCGCAAA ------.(((((.....---.....-........((((((-(....)))))))(((((.((((....(((((.((....)).)))))....))))..))))).....))))). ( -30.00, z-score = -2.88, R) >consensus ______AGUAGAUCUCC__GUCUAUAUCCAGCUAAAUAUA_UUUCCAUGCCGCAAAUGCUGCAAUUUCGAGCC____AAAACGCUCGCAUGUGCAAUCAUUUUUUAUCGCAAA ...............................................(((.((....))((((....(((((..........)))))....)))).............))).. (-11.86 = -11.55 + -0.31)

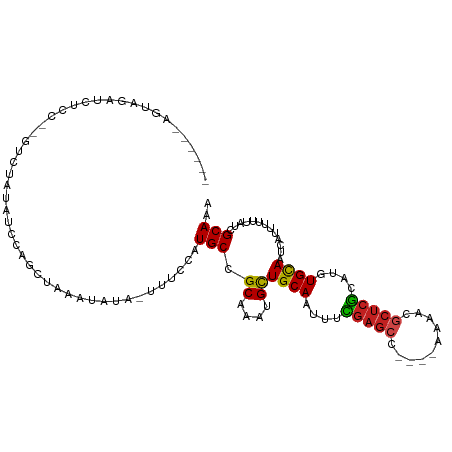

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:54:17 2011