
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 25,184,570 – 25,184,669 |
| Length | 99 |
| Max. P | 0.985924 |

| Location | 25,184,570 – 25,184,669 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 70.10 |
| Shannon entropy | 0.56922 |
| G+C content | 0.48266 |
| Mean single sequence MFE | -25.22 |
| Consensus MFE | -16.93 |
| Energy contribution | -17.43 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.985924 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
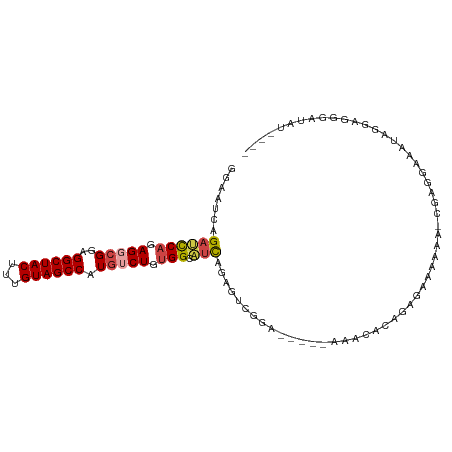
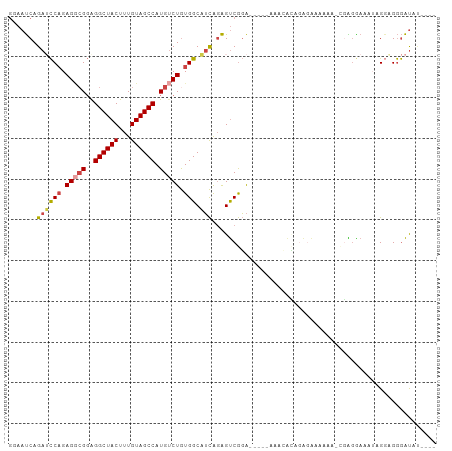
>dm3.chr3R 25184570 99 + 27905053 GGAAUCAGAGCCAGAGGCGGAGGCUACUUUGUAGCCAUGUCUGUGGCAUCAGAGUCGGAAAACACAGGCGAAAAAAAAAAACGAGGAAAUAGAGGGGAU------ ....((.((((((.(((((..((((((...)))))).))))).)))).))...(((..........)))))............................------ ( -26.10, z-score = -2.32, R) >droSim1.chr3R 24859516 98 + 27517382 GGAAUCAGAUCCAGAGGCGGAGGCUACUUUGUAGCCAUGUCUGUGGCAUCAGAGUCGGA-----AAACACAGGGGAAAAAACGAGGAAAUAGGGGGGAUAUAU-- .((.((.((((((.(((((..((((((...)))))).))))).))).))).)).))...-----.......................................-- ( -23.90, z-score = -1.39, R) >droSec1.super_4 4044740 97 + 6179234 GGAAUCAGAUCCAGAGGCGGAGGCUACUUUGUAGCCAUGUCUGUGGCAUCAGAGUCGGA-----AAACACAGGGGAAAAA-CGAGGAAAUAGGGGGGAUACAU-- .((.((.((((((.(((((..((((((...)))))).))))).))).))).)).))...-----................-......................-- ( -23.90, z-score = -1.21, R) >droYak2.chr3R 7529931 103 - 28832112 GCAAUCAGAUCCAGAGGCGGAGGCUACUUUGUAGCCAUGUCUGUGGCAUCAGAGUCGGAAACAAAAAAAUAGAGGAAAAG--AGGAAAUAGGGAGGGAUUUUCCU .......((((((.(((((..((((((...)))))).))))).))).)))......(....)..........(((((((.--................))))))) ( -28.33, z-score = -1.82, R) >droEre2.scaffold_4820 7631109 92 - 10470090 GCAAUCAGAUCCAGAG--GGGGGCUACUUUGUAGCCAUGUCUGUGGCAUCAGAGUCGGA-----AAACACAGGGAAAACG--GAGGAAAUGGGAGGGAUAU---- ...(((...(((....--...((((((...)))))).(.((((((...((.......))-----...)))))).).....--.........)))..)))..---- ( -21.60, z-score = -0.37, R) >droAna3.scaffold_12911 4876822 98 + 5364042 ----CCAGAGUCUGAGACGCAGGCUACUUUGUAGCCAUGACUAUGGCUAUGUGGCCAUGGCAAAAUGGGAAUAAAAAAGUUUUGAGAAGGAGUAGAAGCGUU--- ----...........(((((.((((((...((((((((....))))))))))))))....((((((............)))))).............)))))--- ( -27.50, z-score = -1.82, R) >consensus GGAAUCAGAUCCAGAGGCGGAGGCUACUUUGUAGCCAUGUCUGUGGCAUCAGAGUCGGA_____AAACACAGAGAAAAAA_CGAGGAAAUAGGAGGGAUAU____ .......((((((.(((((..((((((...)))))).))))).))).)))....................................................... (-16.93 = -17.43 + 0.50)

| Location | 25,184,570 – 25,184,669 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 70.10 |
| Shannon entropy | 0.56922 |
| G+C content | 0.48266 |
| Mean single sequence MFE | -17.66 |
| Consensus MFE | -10.54 |
| Energy contribution | -11.43 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.919768 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
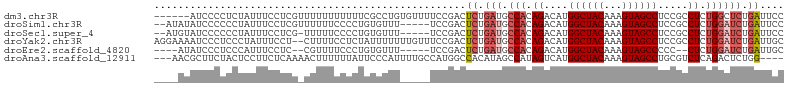
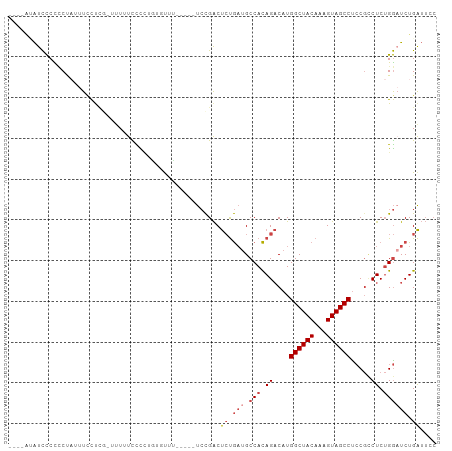
>dm3.chr3R 25184570 99 - 27905053 ------AUCCCCUCUAUUUCCUCGUUUUUUUUUUUCGCCUGUGUUUUCCGACUCUGAUGCCACAGACAUGGCUACAAAGUAGCCUCCGCCUCUGGCUCUGAUUCC ------...........................................((.((.((.((((.((.(..((((((...))))))...).)).)))))).)).)). ( -18.30, z-score = -1.57, R) >droSim1.chr3R 24859516 98 - 27517382 --AUAUAUCCCCCCUAUUUCCUCGUUUUUUCCCCUGUGUUU-----UCCGACUCUGAUGCCACAGACAUGGCUACAAAGUAGCCUCCGCCUCUGGAUCUGAUUCC --.......................................-----...((.((.(((.(((.((.(..((((((...))))))...).)).)))))).)).)). ( -16.30, z-score = -1.08, R) >droSec1.super_4 4044740 97 - 6179234 --AUGUAUCCCCCCUAUUUCCUCG-UUUUUCCCCUGUGUUU-----UCCGACUCUGAUGCCACAGACAUGGCUACAAAGUAGCCUCCGCCUCUGGAUCUGAUUCC --......................-................-----...((.((.(((.(((.((.(..((((((...))))))...).)).)))))).)).)). ( -16.30, z-score = -0.76, R) >droYak2.chr3R 7529931 103 + 28832112 AGGAAAAUCCCUCCCUAUUUCCU--CUUUUCCUCUAUUUUUUUGUUUCCGACUCUGAUGCCACAGACAUGGCUACAAAGUAGCCUCCGCCUCUGGAUCUGAUUGC (((((((................--.)))))))...............(((.((.(((.(((.((.(..((((((...))))))...).)).)))))).))))). ( -18.53, z-score = -0.58, R) >droEre2.scaffold_4820 7631109 92 + 10470090 ----AUAUCCCUCCCAUUUCCUC--CGUUUUCCCUGUGUUU-----UCCGACUCUGAUGCCACAGACAUGGCUACAAAGUAGCCCCC--CUCUGGAUCUGAUUGC ----..........((..(((..--........(((((...-----((.......))...)))))....((((((...))))))...--....)))..))..... ( -17.00, z-score = -1.23, R) >droAna3.scaffold_12911 4876822 98 - 5364042 ---AACGCUUCUACUCCUUCUCAAAACUUUUUUAUUCCCAUUUUGCCAUGGCCACAUAGCCAUAGUCAUGGCUACAAAGUAGCCUGCGUCUCAGACUCUGG---- ---.((((..(((((......................((((......)))).....(((((((....)))))))...)))))...))))............---- ( -19.50, z-score = -1.75, R) >consensus ____AUAUCCCCCCUAUUUCCUCG_UUUUUCCCCUGUGUUU_____UCCGACUCUGAUGCCACAGACAUGGCUACAAAGUAGCCUCCGCCUCUGGAUCUGAUUCC ....................................................((.(((.(((.((....((((((...)))))).....)).)))))).)).... (-10.54 = -11.43 + 0.89)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:54:13 2011