
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 25,184,162 – 25,184,260 |
| Length | 98 |
| Max. P | 0.619871 |

| Location | 25,184,162 – 25,184,260 |
|---|---|
| Length | 98 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 66.05 |
| Shannon entropy | 0.60935 |
| G+C content | 0.36289 |
| Mean single sequence MFE | -22.44 |
| Consensus MFE | -6.21 |
| Energy contribution | -5.35 |
| Covariance contribution | -0.86 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.28 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.619871 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
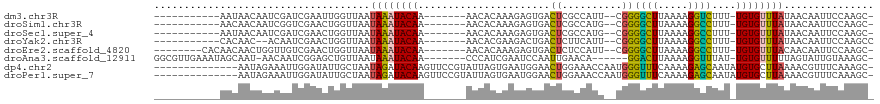
>dm3.chr3R 25184162 98 + 27905053 -----------AAUAACAAUCGAUCGAAUUGGUUAAUAAAUACAA-------AACACAAAGAGUGACUCGCCAUU--CGGGGCUUAAAAGGUCUUU-UGUGUUUAUAACAAUUCCAAGC- -----------..............((((((....((((((((((-------(.......(((((......))))--).(((((.....)))))))-)))))))))..)))))).....- ( -22.20, z-score = -2.64, R) >droSim1.chr3R 24859107 98 + 27517382 -----------AACAACAAUCGGUCGAACUGGUUAAUAAAUACAA-------AACACAAAGAGUGACUCGCCAUG--CGGGGCUUAAAAGGCCUUU-UGUGUUUAUAACAAUUCCAAGC- -----------......((((((.....)))))).((((((((((-------(((((.....)))..((((...)--)))((((.....)))))))-))))))))).............- ( -22.10, z-score = -1.45, R) >droSec1.super_4 4044330 98 + 6179234 -----------AAUAACAAUCGAUCGAACUGGUUAAUAAAUACAA-------AACACAAAGAGUGACUCGCCAUG--CGGGGCUUAAAAGGCCUUU-UGUGUUUAUAACAAUUCCAAGC- -----------..............(((...((((.(((((((((-------(((((.....)))..((((...)--)))((((.....)))))))-))))))))))))..))).....- ( -20.60, z-score = -1.47, R) >droYak2.chr3R 7529523 97 - 28832112 -----------CACAAC--ACAAUCGAACUGGUUAAUAAAUACAA-------AACACGAAGACUGACUCUUCAUU--CGGGGCUUAAAAGGCCUUU-UGUGUUUAUAACAAUUCCAAGCC -----------......--......(((...((((.(((((((((-------(....(((((.....)))))...--..(((((.....)))))))-))))))))))))..)))...... ( -22.00, z-score = -3.02, R) >droEre2.scaffold_4820 7630687 101 - 10470090 --------CACAACAACUGGUUGUCGAACUGGUUAAUAAAUACAA-------AACACAAAGAGUGACUCUCCAUU--CGGGGCUUAAAAGGCCUUU-UGUGUUUACAACAAUUCCAAGC- --------......(((..(((....)))..)))..........(-------(((((((((((((......))))--).(((((.....)))))))-)))))))...............- ( -25.00, z-score = -2.15, R) >droAna3.scaffold_12911 4876342 104 + 5364042 GGCGUUGAAAUAGCAAU-AACAAUCGGAGCUGUUAAUAAAUACAA-------CCCAUCGAAUCCAAUUGAACA------GGACUUAAAAGGUUUAU-UGUGUUUUUAGUAUUGUAAAGC- .((((((......))))-.(((((....((((..((((((((.((-------((..(..(.(((.........------))).)..)..)))))))-).))))..)))))))))...))- ( -13.40, z-score = 1.28, R) >dp4.chr2 2040862 105 + 30794189 --------------AAUAGAAAUUGGAUAUUGCUAAUAGAUACAAGUUCCGUAUUAGUGAAUGGAACUGGAAACCAAUGGGUUUCAAAAGAGCAAUAUGUGCUUAAAACGUUUCAAAGC- --------------....(((((...((((((((..........((((((((........)))))))).((((((....)))))).....))))))))((.......))))))).....- ( -27.10, z-score = -3.56, R) >droPer1.super_7 2208560 105 + 4445127 --------------AAUAGAAAUUGGAUAUUGCUAAUAGAUACAAGUUCCGUAUUAGUGAAUGGAACUGGAAACCAAUGGGUUUCAAAAGAGCAAUAUGUGCUUAAAACGUUUCAAAGC- --------------....(((((...((((((((..........((((((((........)))))))).((((((....)))))).....))))))))((.......))))))).....- ( -27.10, z-score = -3.56, R) >consensus ___________AACAACAAACAAUCGAACUGGUUAAUAAAUACAA_______AACACAAAGAGUGACUCGCCAUU__CGGGGCUUAAAAGGCCUUU_UGUGUUUAUAACAAUUCCAAGC_ ....................................((((((((......................((..........))((((.....))))....))))))))............... ( -6.21 = -5.35 + -0.86)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:54:12 2011